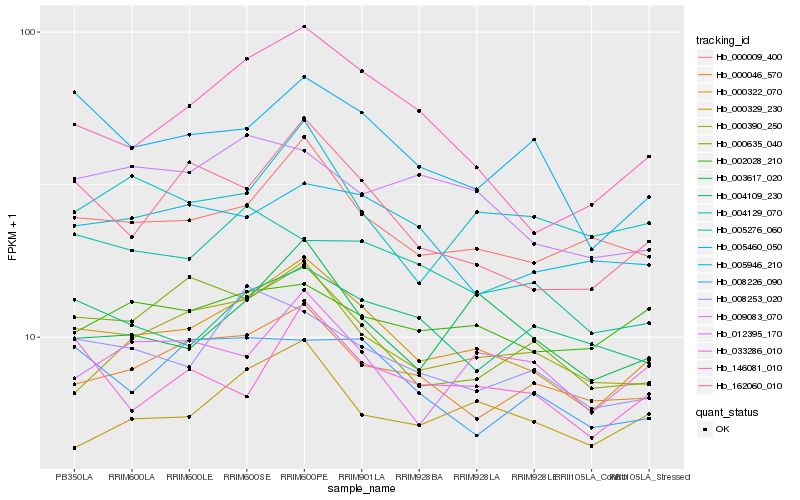
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000322_070 |
0.0 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_000009_400 |
0.0679434822 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 3 |
Hb_000046_570 |
0.0766218781 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 4 |
Hb_162060_010 |
0.0784861909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003617_020 |
0.0786061479 |
- |
- |
PREDICTED: uncharacterized protein LOC105638179 [Jatropha curcas] |
| 6 |
Hb_008253_020 |
0.0790791541 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 7 |
Hb_000390_250 |
0.0811092008 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 8 |
Hb_005946_210 |
0.0813211725 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 9 |
Hb_000329_230 |
0.0819555967 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 10 |
Hb_146081_010 |
0.0829105946 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 11 |
Hb_012395_170 |
0.0851325126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000635_040 |
0.087062706 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 13 |
Hb_005276_060 |
0.0881903183 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_033286_010 |
0.088802996 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 15 |
Hb_005460_050 |
0.089214688 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 16 |
Hb_009083_070 |
0.0896928211 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 17 |
Hb_004109_230 |
0.0899225906 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_002028_210 |
0.090549648 |
- |
- |
PREDICTED: uncharacterized protein LOC105638038 [Jatropha curcas] |
| 19 |
Hb_004129_070 |
0.0912118816 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 20 |
Hb_008226_090 |
0.0914720743 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |