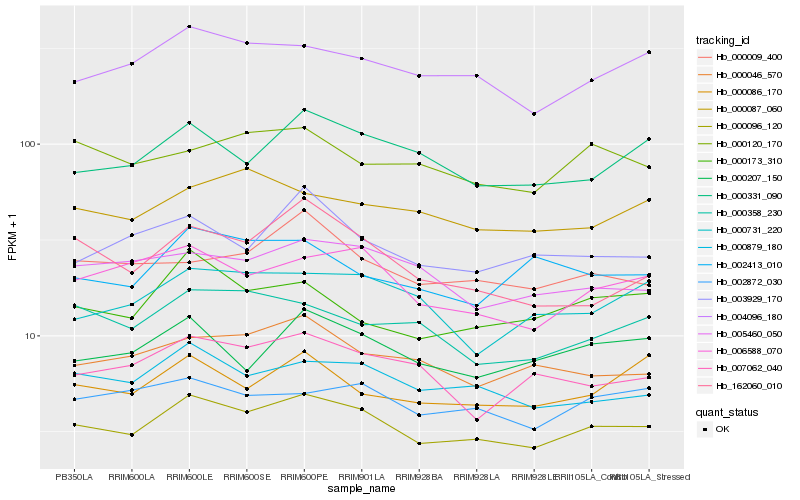
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000096_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 2 |
Hb_000879_180 |
0.0655577175 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 3 |
Hb_004096_180 |
0.0709521977 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 4 |
Hb_006588_070 |
0.0740412039 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 5 |
Hb_000331_090 |
0.0794151908 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 6 |
Hb_000207_150 |
0.0806149582 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 7 |
Hb_000046_570 |
0.0817497409 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 8 |
Hb_002872_030 |
0.081982571 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 9 |
Hb_005460_050 |
0.0832035975 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 10 |
Hb_000009_400 |
0.0841158054 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 11 |
Hb_003929_170 |
0.0849360284 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 12 |
Hb_000173_310 |
0.0862285081 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007062_040 |
0.0879111166 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 14 |
Hb_000087_060 |
0.0879501556 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 15 |
Hb_000358_230 |
0.0884026573 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 16 |
Hb_000120_170 |
0.0888382162 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 17 |
Hb_000086_170 |
0.088935997 |
- |
- |
PREDICTED: serine/threonine-protein kinase 19 [Jatropha curcas] |
| 18 |
Hb_162060_010 |
0.0890678975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002413_010 |
0.0904256618 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_000731_220 |
0.0907115593 |
- |
- |
PREDICTED: derlin-2.2 [Jatropha curcas] |