| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
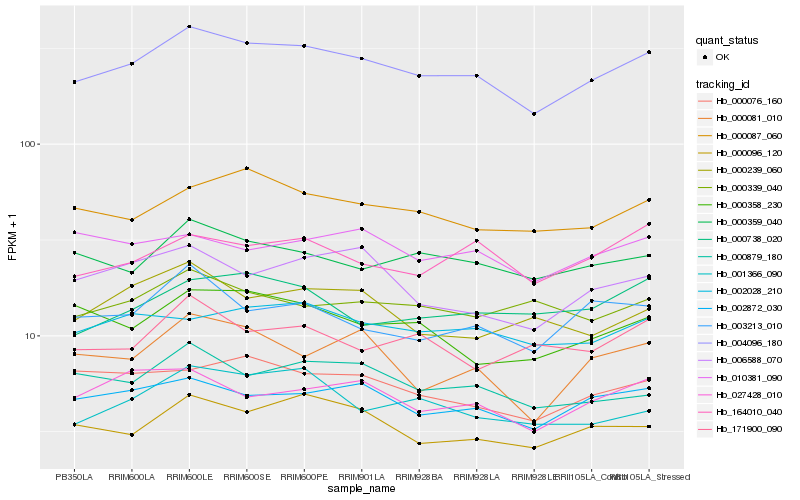
Hb_004096_180 |
0.0 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 2 |
Hb_002872_030 |
0.0628046008 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 3 |
Hb_000087_060 |
0.0676338918 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 4 |
Hb_000096_120 |
0.0709521977 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 5 |
Hb_002028_210 |
0.0710007795 |
- |
- |
PREDICTED: uncharacterized protein LOC105638038 [Jatropha curcas] |
| 6 |
Hb_000359_040 |
0.071142789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_027428_010 |
0.0742233278 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000879_180 |
0.0742308967 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 9 |
Hb_000076_160 |
0.0748576388 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 10 |
Hb_164010_040 |
0.0769630692 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 11 |
Hb_001366_090 |
0.0780131258 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 12 |
Hb_003213_010 |
0.0785136453 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 13 |
Hb_006588_070 |
0.0788538787 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 14 |
Hb_000358_230 |
0.0797599281 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 15 |
Hb_000738_020 |
0.0811441162 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000339_040 |
0.0821305423 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_171900_090 |
0.0842491393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000081_010 |
0.0845760724 |
- |
- |
hypothetical protein B456_012G056000 [Gossypium raimondii] |
| 19 |
Hb_000239_060 |
0.0849475897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_010381_090 |
0.0851186201 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |