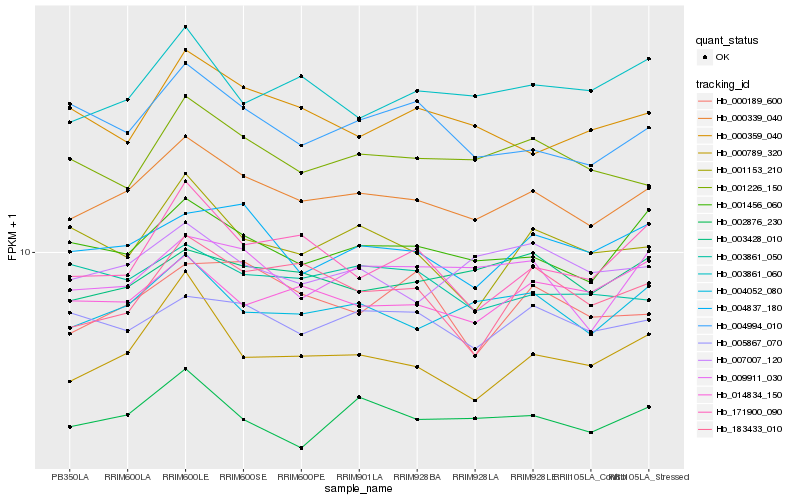
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000339_040 |
0.0 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_004052_080 |
0.0448139514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001456_060 |
0.048899009 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 4 |
Hb_002876_230 |
0.052212756 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 5 |
Hb_009911_030 |
0.0553606753 |
transcription factor |
TF Family: FAR1 |
far-red impaired response 1 -like protein [Gossypium arboreum] |
| 6 |
Hb_004837_180 |
0.0555548767 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_005867_070 |
0.0559340653 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_003428_010 |
0.056862657 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 9 |
Hb_001226_150 |
0.0579252749 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 10 |
Hb_000789_320 |
0.058892091 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_007007_120 |
0.0593943786 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004994_010 |
0.0602894339 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 13 |
Hb_003861_060 |
0.0609926887 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 14 |
Hb_003861_050 |
0.0619463351 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 15 |
Hb_014834_150 |
0.0622320307 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_171900_090 |
0.0641640209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_183433_010 |
0.0643155049 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 18 |
Hb_000359_040 |
0.0648793934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001153_210 |
0.0659471626 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000189_600 |
0.0667392995 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |