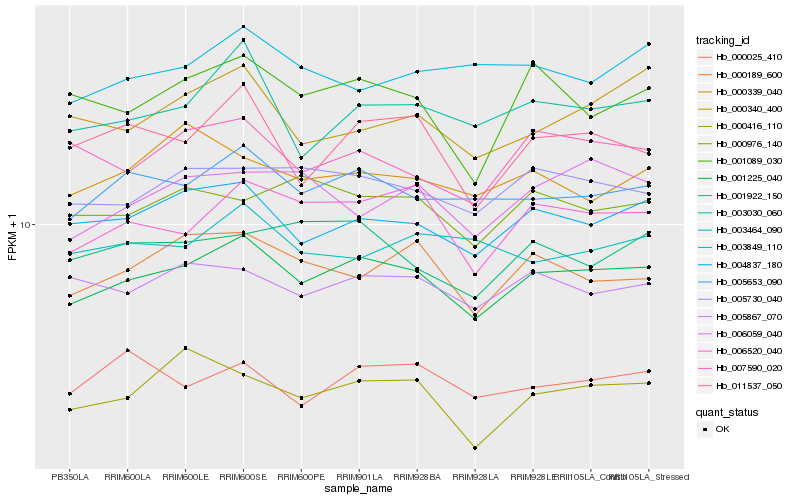
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001225_040 |
0.0 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 2 |
Hb_004837_180 |
0.051324197 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_005653_090 |
0.0528227788 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 4 |
Hb_006520_040 |
0.0546146948 |
- |
- |
PREDICTED: uncharacterized protein LOC105641714 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005730_040 |
0.0591048195 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 6 |
Hb_003030_060 |
0.0628421601 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 7 |
Hb_011537_050 |
0.0640729014 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005867_070 |
0.0641931759 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_000189_600 |
0.0657993005 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_007590_020 |
0.0659279572 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 11 |
Hb_001089_030 |
0.0660039446 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 12 |
Hb_000976_140 |
0.0713539847 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001922_150 |
0.0714680401 |
- |
- |
PREDICTED: WD repeat-containing protein 91 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_006059_040 |
0.074226326 |
transcription factor |
TF Family: SOH1 |
hypothetical protein PHAVU_007G063000g [Phaseolus vulgaris] |
| 15 |
Hb_000340_400 |
0.0747372107 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 16 |
Hb_003849_110 |
0.0756278477 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 17 |
Hb_003464_090 |
0.0766499969 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 18 |
Hb_000025_410 |
0.0767433499 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g01110-like [Populus euphratica] |
| 19 |
Hb_000339_040 |
0.0776466351 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 20 |
Hb_000416_110 |
0.0777269192 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |