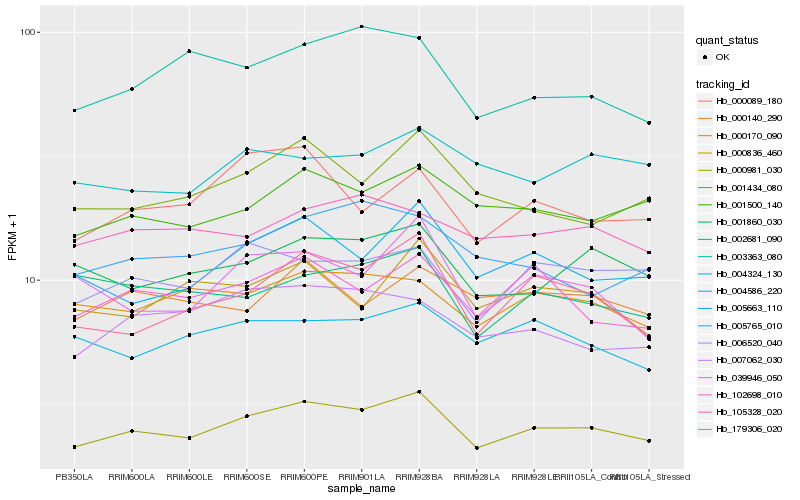
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000981_030 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 2 |
Hb_001500_140 |
0.0638088515 |
- |
- |
pelota, putative [Ricinus communis] |
| 3 |
Hb_179306_020 |
0.0649209094 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001860_030 |
0.0668208334 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 5 |
Hb_006520_040 |
0.0673256424 |
- |
- |
PREDICTED: uncharacterized protein LOC105641714 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005663_110 |
0.0693557884 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000170_090 |
0.0693903096 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 8 |
Hb_102698_010 |
0.0702903186 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001434_080 |
0.0725860563 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 10 |
Hb_004586_220 |
0.0749955553 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 11 |
Hb_007062_030 |
0.0753192832 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 12 |
Hb_105328_020 |
0.0753537654 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 13 |
Hb_003363_080 |
0.0770681216 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 14 |
Hb_005765_010 |
0.0790287129 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004324_130 |
0.0796674894 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 16 |
Hb_000836_460 |
0.0798335697 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |
| 17 |
Hb_002681_090 |
0.0798809396 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000089_180 |
0.0800044689 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 19 |
Hb_000140_290 |
0.0803236789 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 59 kDa protein isoform X1 [Populus euphratica] |
| 20 |
Hb_039946_050 |
0.0820989567 |
- |
- |
catalytic, putative [Ricinus communis] |