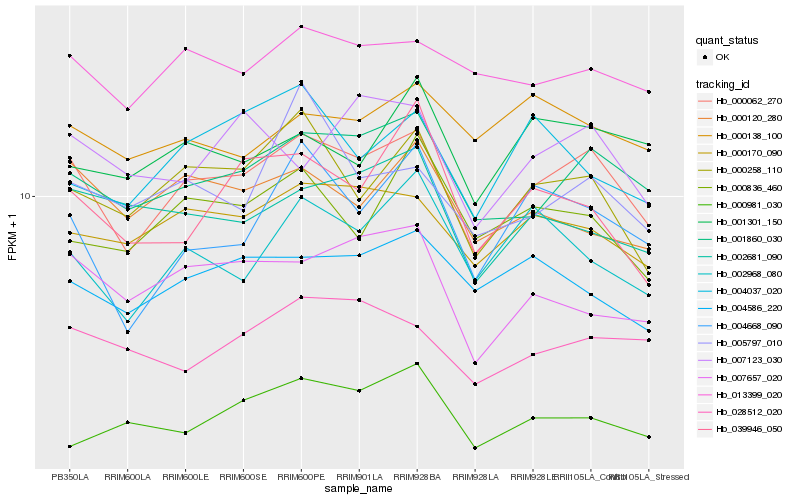
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_270 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001860_030 |
0.0588280531 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 3 |
Hb_000170_090 |
0.0706475107 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 4 |
Hb_000258_110 |
0.0726406036 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 5 |
Hb_013399_020 |
0.075990204 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 6 |
Hb_004668_090 |
0.0817650661 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 7 |
Hb_004586_220 |
0.0819967831 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 8 |
Hb_007657_020 |
0.0826282672 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 9 |
Hb_000120_280 |
0.0842872793 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 10 |
Hb_028512_020 |
0.0846430464 |
- |
- |
PREDICTED: protein S-acyltransferase 11 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002681_090 |
0.0852280532 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002968_080 |
0.0855585275 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 13 |
Hb_007123_030 |
0.0872893848 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 14 |
Hb_001301_150 |
0.0889216065 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 15 |
Hb_000138_100 |
0.0889346006 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 16 |
Hb_000836_460 |
0.0901818356 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |
| 17 |
Hb_004037_020 |
0.0911478082 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005797_010 |
0.0917994951 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_039946_050 |
0.0918178823 |
- |
- |
catalytic, putative [Ricinus communis] |
| 20 |
Hb_000981_030 |
0.0921610329 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |