| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
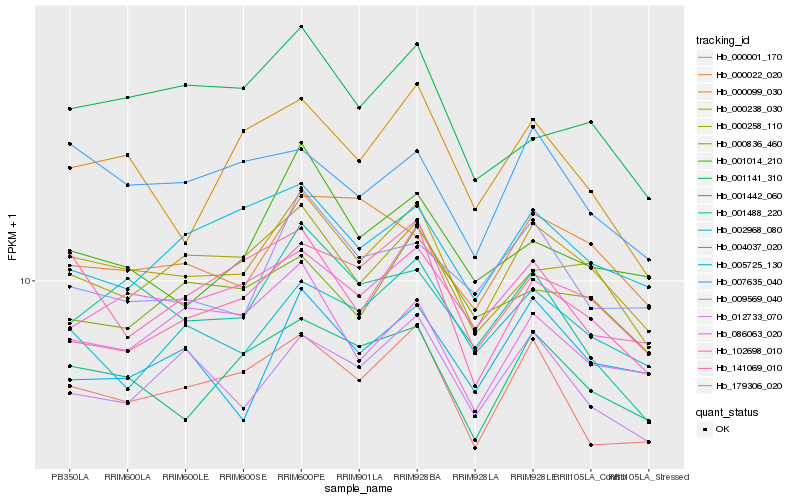
Hb_000238_030 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_001442_060 |
0.0628460332 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000258_110 |
0.0637221542 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000001_170 |
0.0703543866 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_102698_010 |
0.0710942202 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_012733_070 |
0.0772690356 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 7 |
Hb_086063_020 |
0.0781287553 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 8 |
Hb_002968_080 |
0.0788428445 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |
| 9 |
Hb_004037_020 |
0.0803068118 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007635_040 |
0.0806347477 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 11 |
Hb_179306_020 |
0.0811193912 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009569_040 |
0.0825322583 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 13 |
Hb_001014_210 |
0.0834401885 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 14 |
Hb_000099_030 |
0.0838297052 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_141069_010 |
0.0841229214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001488_220 |
0.0846222422 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 17 |
Hb_000836_460 |
0.084656641 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |
| 18 |
Hb_001141_310 |
0.0849531327 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 19 |
Hb_000022_020 |
0.0854935836 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 20 |
Hb_005725_130 |
0.086566799 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |