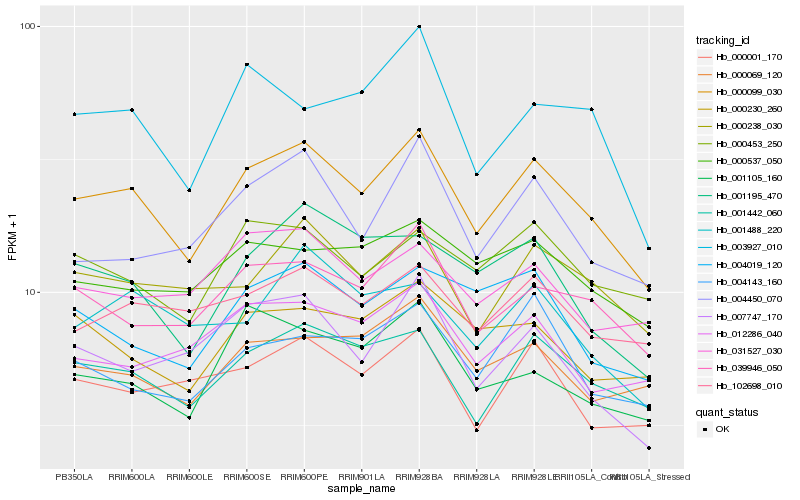
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000099_030 |
0.0 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_001442_060 |
0.0688187181 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 3 |
Hb_039946_050 |
0.0767487634 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_000238_030 |
0.0838297052 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_102698_010 |
0.0888869622 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000453_250 |
0.0889791746 |
- |
- |
PREDICTED: uncharacterized protein LOC105637185 [Jatropha curcas] |
| 7 |
Hb_000001_170 |
0.0900360554 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_001195_470 |
0.090470134 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 9 |
Hb_000069_120 |
0.0909533252 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 10 |
Hb_004450_070 |
0.091142573 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 11 |
Hb_004019_120 |
0.0927430725 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 12 |
Hb_000537_050 |
0.0933686281 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 13 |
Hb_007747_170 |
0.0934790527 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 14 |
Hb_004143_160 |
0.0937677226 |
- |
- |
SAB, putative [Ricinus communis] |
| 15 |
Hb_031527_030 |
0.094449633 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 16 |
Hb_001105_160 |
0.0952762334 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 17 |
Hb_001488_220 |
0.0959112753 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 18 |
Hb_000230_260 |
0.0959136965 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 19 |
Hb_012286_040 |
0.0960390443 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 20 |
Hb_003927_010 |
0.0962909066 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |