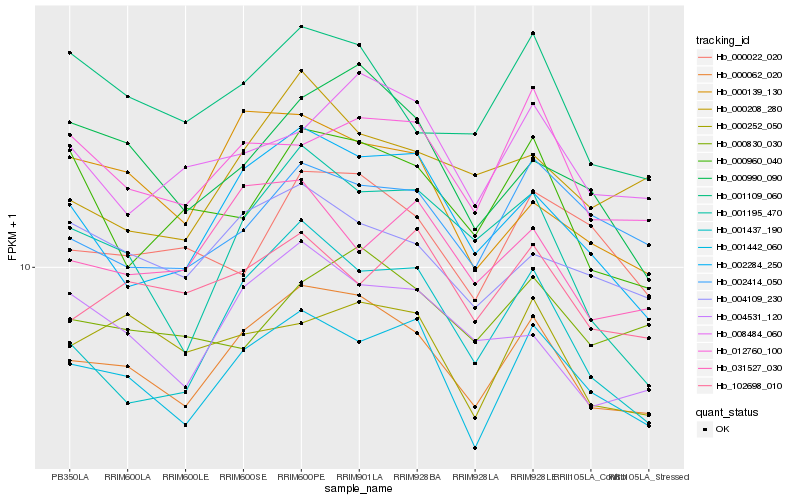
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_020 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_001442_060 |
0.0607841401 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002284_250 |
0.0732677428 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 4 |
Hb_004109_230 |
0.0778819602 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_004531_120 |
0.0784255892 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 6 |
Hb_001109_060 |
0.0798803051 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_012760_100 |
0.0799479485 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 8 |
Hb_000252_050 |
0.0840228206 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 9 |
Hb_001437_190 |
0.0860930284 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_000990_090 |
0.0863446078 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_002414_050 |
0.0864965278 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 12 |
Hb_008484_060 |
0.087040514 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 13 |
Hb_000139_130 |
0.0871298462 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 14 |
Hb_031527_030 |
0.0888003559 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 15 |
Hb_000022_020 |
0.0893209084 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 16 |
Hb_001195_470 |
0.0897224159 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 17 |
Hb_000208_280 |
0.0905420464 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 18 |
Hb_000960_040 |
0.0909864122 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 19 |
Hb_000830_030 |
0.0911524661 |
- |
- |
PREDICTED: uncharacterized protein LOC105644214 [Jatropha curcas] |
| 20 |
Hb_102698_010 |
0.0912094296 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |