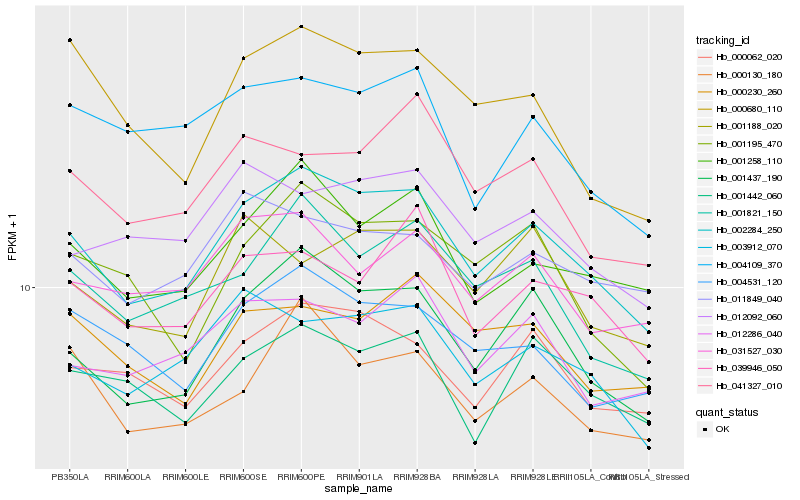
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_250 |
0.0 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 2 |
Hb_001437_190 |
0.0552231525 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_001258_110 |
0.0685798159 |
- |
- |
transporter, putative [Ricinus communis] |
| 4 |
Hb_000062_020 |
0.0732677428 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_001821_150 |
0.0745584634 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 6 |
Hb_003912_070 |
0.0785544438 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 7 |
Hb_004531_120 |
0.0803799123 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 8 |
Hb_000130_180 |
0.0810978053 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001442_060 |
0.0834422575 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 10 |
Hb_039946_050 |
0.0846128253 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_012286_040 |
0.0851428509 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 12 |
Hb_031527_030 |
0.0862180348 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 13 |
Hb_001195_470 |
0.0866522924 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 14 |
Hb_011849_040 |
0.0868078509 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_041327_010 |
0.0880187884 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 16 |
Hb_000680_110 |
0.0891909579 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 17 |
Hb_012092_060 |
0.089307365 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 18 |
Hb_004109_370 |
0.0902588099 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 19 |
Hb_000230_260 |
0.0909690523 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 20 |
Hb_001188_020 |
0.0910419154 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |