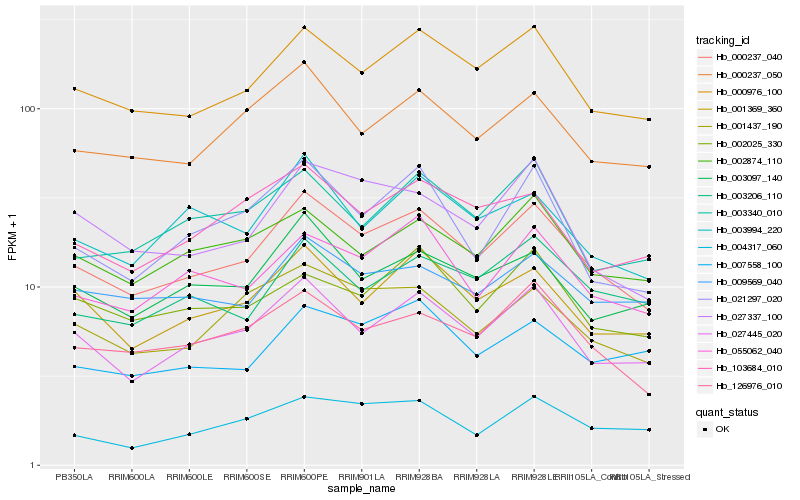
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000237_040 |
0.0 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 2 |
Hb_000976_100 |
0.0596401967 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 3 |
Hb_027445_020 |
0.0648822196 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 4 |
Hb_126976_010 |
0.0756676225 |
- |
- |
- |
| 5 |
Hb_001369_360 |
0.079988682 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 6 |
Hb_000237_050 |
0.0813337597 |
- |
- |
CP2 [Hevea brasiliensis] |
| 7 |
Hb_055062_040 |
0.0844809338 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 8 |
Hb_001437_190 |
0.0845925689 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_003097_140 |
0.090713288 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 10 |
Hb_027337_100 |
0.0913019232 |
- |
- |
Putative phagocytic receptor 1b [Gossypium arboreum] |
| 11 |
Hb_002874_110 |
0.0919331975 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 12 |
Hb_003994_220 |
0.0933996306 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 13 |
Hb_021297_020 |
0.0936256671 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 14 |
Hb_003206_110 |
0.0947759383 |
- |
- |
Putative 1-aminocyclopropane-1-carboxylate deaminase [Gossypium arboreum] |
| 15 |
Hb_009569_040 |
0.0969938788 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 16 |
Hb_103684_010 |
0.0979727074 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |
| 17 |
Hb_004317_060 |
0.0982463966 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 18 |
Hb_002025_330 |
0.0991150818 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 19 |
Hb_003340_010 |
0.0992784646 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 20 |
Hb_007558_100 |
0.1000934817 |
- |
- |
PREDICTED: uncharacterized protein LOC105649092 isoform X2 [Jatropha curcas] |