| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
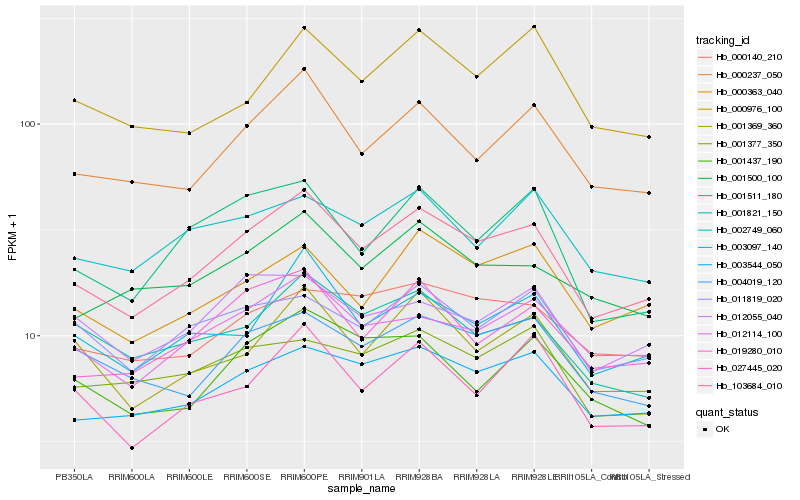
Hb_103684_010 |
0.0 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |
| 2 |
Hb_000237_050 |
0.0678757454 |
- |
- |
CP2 [Hevea brasiliensis] |
| 3 |
Hb_003544_050 |
0.0685465182 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 4 |
Hb_019280_010 |
0.0715206759 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003097_140 |
0.0772970254 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 6 |
Hb_012114_100 |
0.0777327701 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 7 |
Hb_001369_360 |
0.080181304 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 8 |
Hb_000363_040 |
0.0805527505 |
- |
- |
Protein transport protein Sec24A, putative [Ricinus communis] |
| 9 |
Hb_027445_020 |
0.0816189818 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 10 |
Hb_001500_100 |
0.0823083846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004019_120 |
0.0828006396 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 12 |
Hb_001511_180 |
0.0847194679 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |
| 13 |
Hb_000140_210 |
0.0863925129 |
- |
- |
sec10, putative [Ricinus communis] |
| 14 |
Hb_001821_150 |
0.0866606212 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 15 |
Hb_002749_060 |
0.0888458572 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 16 |
Hb_001437_190 |
0.0890130556 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_000976_100 |
0.0905183203 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 18 |
Hb_011819_020 |
0.0905670329 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001377_350 |
0.0911460054 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 20 |
Hb_012055_040 |
0.0925451499 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |