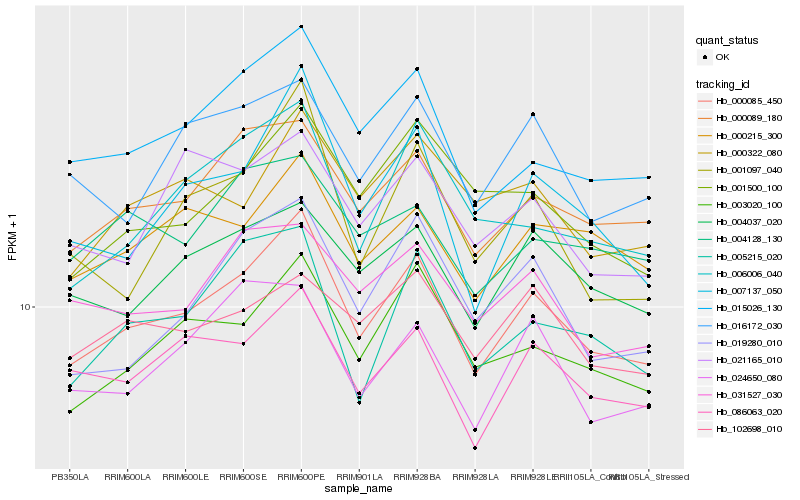
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_450 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 2 |
Hb_015026_130 |
0.0581300255 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 3 |
Hb_000089_180 |
0.05874205 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 4 |
Hb_003020_100 |
0.0620533044 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 5 |
Hb_086063_020 |
0.0664841424 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 6 |
Hb_007137_050 |
0.066790113 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_004037_020 |
0.0688101933 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000215_300 |
0.0722025822 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 9 |
Hb_005215_020 |
0.0737666956 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 10 |
Hb_016172_030 |
0.0749861044 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 11 |
Hb_031527_030 |
0.0758315117 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 12 |
Hb_019280_010 |
0.076158633 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 13 |
Hb_006006_040 |
0.0762241709 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 14 |
Hb_024650_080 |
0.0767836396 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 15 |
Hb_001500_100 |
0.0786936018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_102698_010 |
0.0799442263 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_004128_130 |
0.0799626154 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000322_080 |
0.0803198562 |
- |
- |
hypothetical protein JCGZ_07423 [Jatropha curcas] |
| 19 |
Hb_001097_040 |
0.0803953014 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 20 |
Hb_021165_010 |
0.0821816523 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |