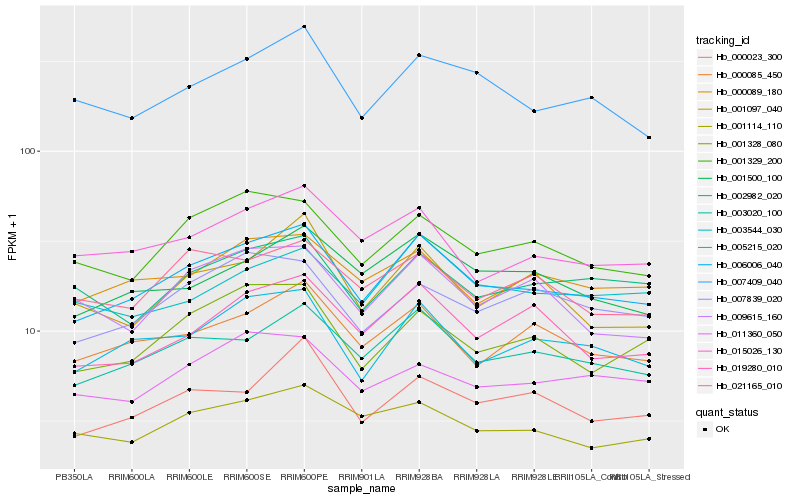
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006006_040 |
0.0 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 2 |
Hb_003020_100 |
0.0574083725 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 3 |
Hb_005215_020 |
0.0649875685 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 4 |
Hb_000085_450 |
0.0762241709 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 5 |
Hb_001329_200 |
0.0771876767 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 6 |
Hb_001328_080 |
0.0781671528 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 7 |
Hb_007409_040 |
0.0790061617 |
- |
- |
actin family protein [Populus trichocarpa] |
| 8 |
Hb_001500_100 |
0.0799938955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_019280_010 |
0.0804778246 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 10 |
Hb_015026_130 |
0.0807952872 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 11 |
Hb_001114_110 |
0.0812044383 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 12 |
Hb_000089_180 |
0.0823360201 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 13 |
Hb_007839_020 |
0.0849273397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000023_300 |
0.0854400213 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 15 |
Hb_021165_010 |
0.0872590287 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003544_030 |
0.0891548665 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 17 |
Hb_009615_160 |
0.0900732651 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 18 |
Hb_001097_040 |
0.0906749315 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 19 |
Hb_011360_050 |
0.0909895677 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002982_020 |
0.0914503619 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |