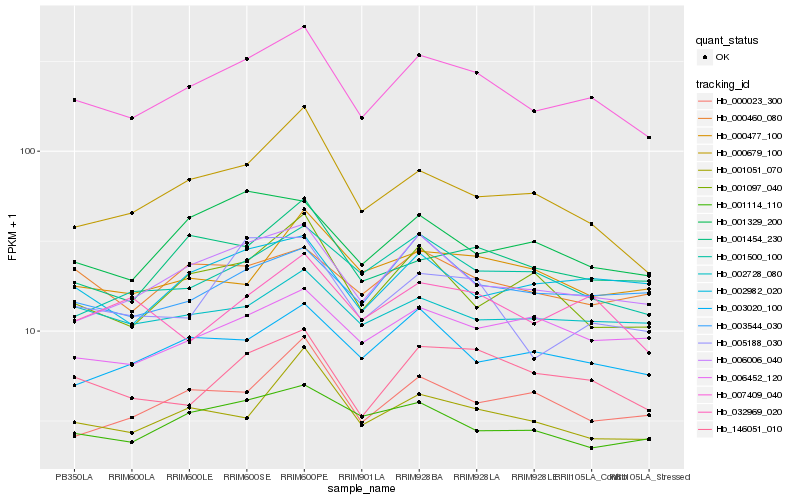
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007409_040 |
0.0 |
- |
- |
actin family protein [Populus trichocarpa] |
| 2 |
Hb_006006_040 |
0.0790061617 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 3 |
Hb_146051_010 |
0.0831166474 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 4 |
Hb_001454_230 |
0.0929380688 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 5 |
Hb_001500_100 |
0.0936477715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001051_070 |
0.0962760553 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001114_110 |
0.0976590725 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 8 |
Hb_003020_100 |
0.0977189267 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 9 |
Hb_002982_020 |
0.097784222 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 10 |
Hb_003544_030 |
0.0988178467 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 11 |
Hb_032969_020 |
0.0998308521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000460_080 |
0.1004809955 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |
| 13 |
Hb_002728_080 |
0.100941762 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_005188_030 |
0.100974456 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000679_100 |
0.1018240797 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001329_200 |
0.1018427344 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 17 |
Hb_006452_120 |
0.1031809108 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 18 |
Hb_001097_040 |
0.1057982872 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 19 |
Hb_000477_100 |
0.1064805145 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 20 |
Hb_000023_300 |
0.1066912141 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |