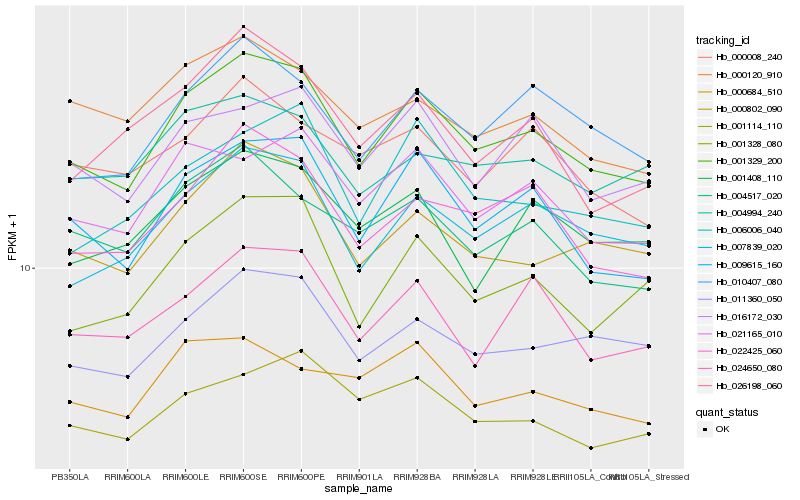
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001329_200 |
0.0 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 2 |
Hb_009615_160 |
0.0522632289 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 3 |
Hb_007839_020 |
0.061044569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_022425_060 |
0.0638372144 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_000802_090 |
0.064372353 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 6 |
Hb_011360_050 |
0.0669845069 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001328_080 |
0.0678794711 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 8 |
Hb_010407_080 |
0.0682226301 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 9 |
Hb_021165_010 |
0.0706127652 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004517_020 |
0.0713914759 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 11 |
Hb_024650_080 |
0.0749369035 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 12 |
Hb_016172_030 |
0.0750895275 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 13 |
Hb_006006_040 |
0.0771876767 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 14 |
Hb_001408_110 |
0.0779317016 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 15 |
Hb_001114_110 |
0.078163197 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 16 |
Hb_026198_060 |
0.0807961882 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 17 |
Hb_000120_910 |
0.0808154737 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 18 |
Hb_004994_240 |
0.0814896755 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 19 |
Hb_000008_240 |
0.0815117206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000684_510 |
0.0821734619 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |