| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
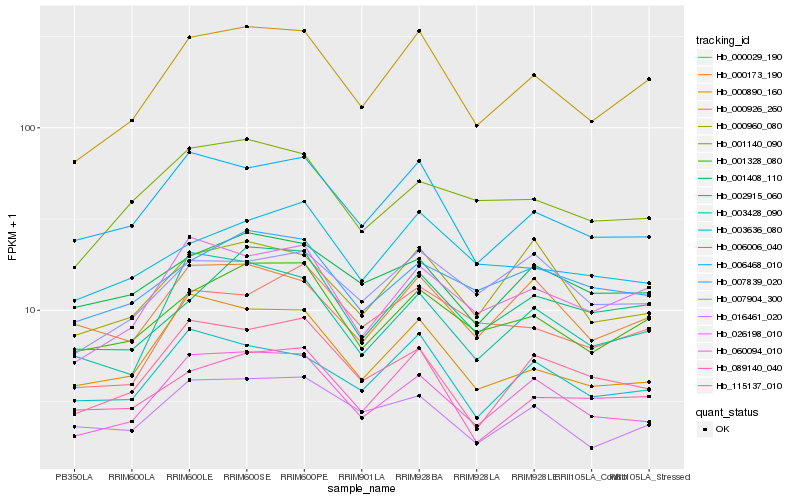
Hb_000926_260 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_026198_010 |
0.0772895574 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001328_080 |
0.080752484 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 4 |
Hb_060094_010 |
0.0826611097 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006468_010 |
0.086676062 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003636_080 |
0.0939997674 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 7 |
Hb_089140_040 |
0.0958606309 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |
| 8 |
Hb_000890_160 |
0.0962043861 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 9 |
Hb_003428_090 |
0.0969083185 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 10 |
Hb_002915_060 |
0.0982941684 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 11 |
Hb_115137_010 |
0.0994821432 |
- |
- |
PREDICTED: probable protein S-acyltransferase 23 isoform X2 [Jatropha curcas] |
| 12 |
Hb_016461_020 |
0.100695492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000029_190 |
0.1018422917 |
- |
- |
hypothetical protein CISIN_1g025338mg [Citrus sinensis] |
| 14 |
Hb_000173_190 |
0.1033176287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000960_080 |
0.1063606676 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_001140_090 |
0.1071922761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007904_300 |
0.1073988005 |
- |
- |
copine, putative [Ricinus communis] |
| 18 |
Hb_006006_040 |
0.1074850562 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 19 |
Hb_001408_110 |
0.1078910141 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 20 |
Hb_007839_020 |
0.1090022865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |