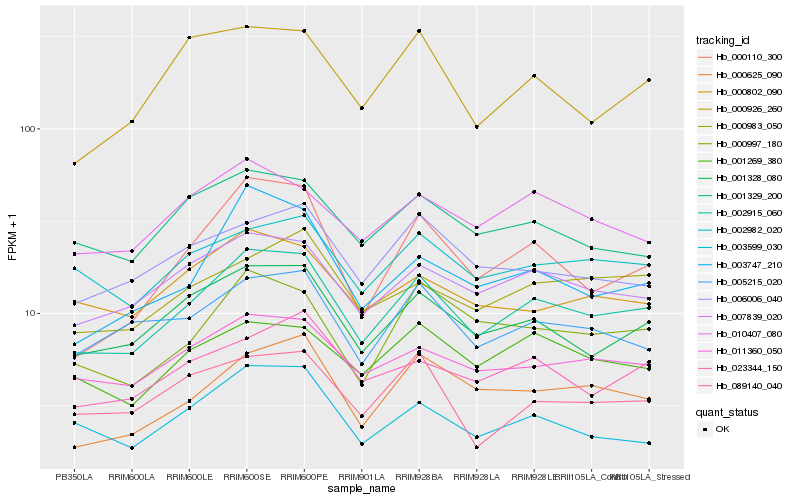
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002915_060 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 2 |
Hb_001328_080 |
0.0702444549 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 3 |
Hb_007839_020 |
0.0730499292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011360_050 |
0.0819726099 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005215_020 |
0.0911372756 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 6 |
Hb_023344_150 |
0.0917239624 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 7 |
Hb_002982_020 |
0.0917393472 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 8 |
Hb_001269_380 |
0.0917397714 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000997_180 |
0.0918683657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001329_200 |
0.092376823 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000983_050 |
0.0929667693 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_000625_090 |
0.0931601548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010407_080 |
0.0936450564 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003747_210 |
0.0948882631 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 15 |
Hb_006006_040 |
0.0956704196 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 16 |
Hb_000110_300 |
0.0956881372 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 17 |
Hb_000802_090 |
0.0976027573 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 18 |
Hb_003599_030 |
0.0981607702 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000926_260 |
0.0982941684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_089140_040 |
0.0988144887 |
- |
- |
PREDICTED: uncharacterized protein LOC105637966 [Jatropha curcas] |