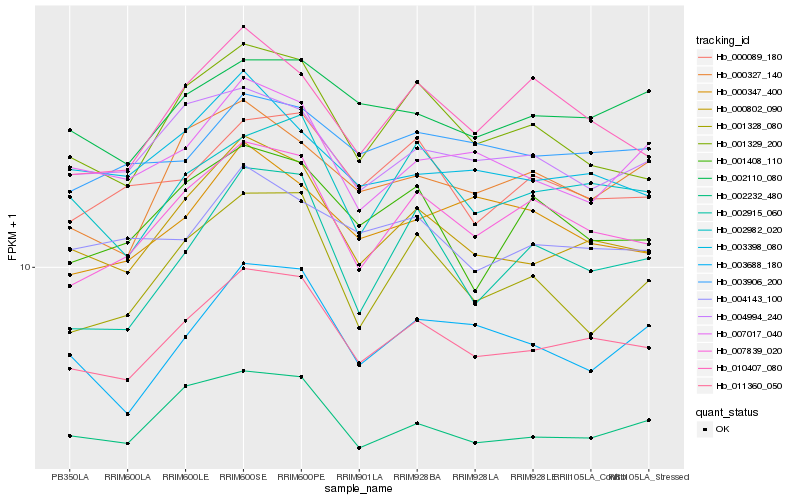
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011360_050 |
0.0 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000802_090 |
0.0426769801 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 3 |
Hb_007839_020 |
0.0568581412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002232_480 |
0.0591010855 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 5 |
Hb_002982_020 |
0.0602576306 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 6 |
Hb_001329_200 |
0.0669845069 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 7 |
Hb_000089_180 |
0.0709639576 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 8 |
Hb_002110_080 |
0.0731294715 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 9 |
Hb_004994_240 |
0.0743507092 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 10 |
Hb_003688_180 |
0.0745829033 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 11 |
Hb_003906_200 |
0.0769437945 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 12 |
Hb_000327_140 |
0.0776419931 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001408_110 |
0.0781994948 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 14 |
Hb_010407_080 |
0.0782843123 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007017_040 |
0.0787824829 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 16 |
Hb_004143_100 |
0.0794497174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001328_080 |
0.0815810651 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 18 |
Hb_003398_080 |
0.0817109292 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 19 |
Hb_002915_060 |
0.0819726099 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 20 |
Hb_000347_400 |
0.0820730017 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |