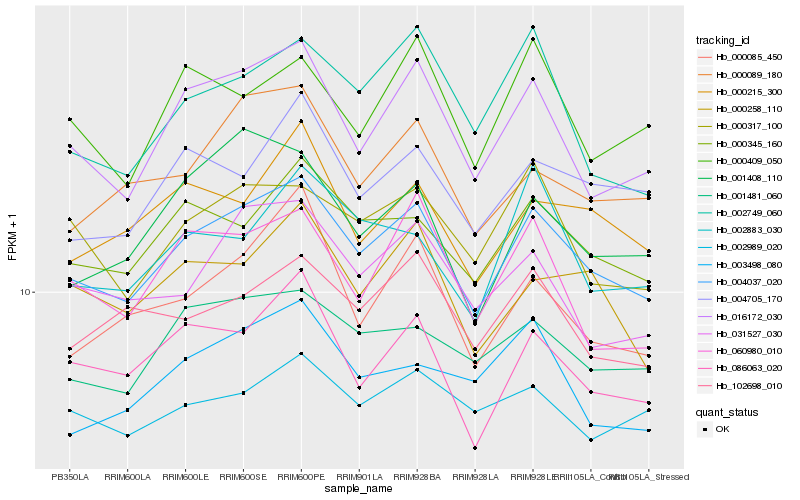
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004037_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 2 |
Hb_016172_030 |
0.0497815828 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 3 |
Hb_000345_160 |
0.0509394344 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 4 |
Hb_001481_060 |
0.0575139321 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_086063_020 |
0.0578406352 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 6 |
Hb_000317_100 |
0.0624507654 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 7 |
Hb_004705_170 |
0.0627207543 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 8 |
Hb_000089_180 |
0.0638202085 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 9 |
Hb_102698_010 |
0.0638353193 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_031527_030 |
0.0644299909 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 11 |
Hb_000215_300 |
0.064546866 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 12 |
Hb_000258_110 |
0.0657380463 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_060980_010 |
0.0687881576 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_000085_450 |
0.0688101933 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 15 |
Hb_001408_110 |
0.0692008041 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 16 |
Hb_002883_030 |
0.0692324697 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 17 |
Hb_000409_050 |
0.0694788667 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 18 |
Hb_003498_080 |
0.0700190833 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_002749_060 |
0.0700866504 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 20 |
Hb_002989_020 |
0.071091435 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |