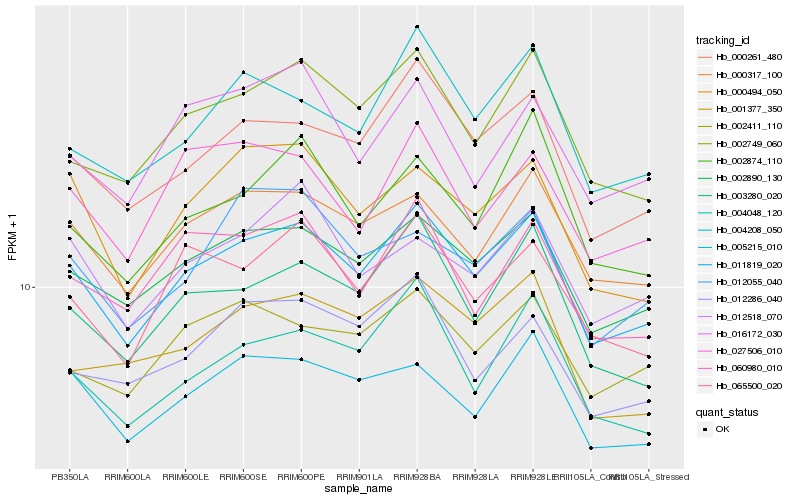
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011819_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002890_130 |
0.0393450143 |
- |
- |
tip120, putative [Ricinus communis] |
| 3 |
Hb_000261_480 |
0.0447091431 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 4 |
Hb_000317_100 |
0.0488061467 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 5 |
Hb_002749_060 |
0.0522806226 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 6 |
Hb_003280_020 |
0.0531021785 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005215_010 |
0.0557661871 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004048_120 |
0.0570442384 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 9 |
Hb_065500_020 |
0.0576176518 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 10 |
Hb_004208_050 |
0.0586070221 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 11 |
Hb_002411_110 |
0.0607865215 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 12 |
Hb_000494_050 |
0.0615300657 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_060980_010 |
0.0640000234 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_012518_070 |
0.064683049 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 15 |
Hb_012286_040 |
0.0648676222 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 16 |
Hb_002874_110 |
0.0650180435 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 17 |
Hb_027506_010 |
0.0660293987 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 18 |
Hb_001377_350 |
0.0666033425 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 19 |
Hb_016172_030 |
0.068498295 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 20 |
Hb_012055_040 |
0.0691512244 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |