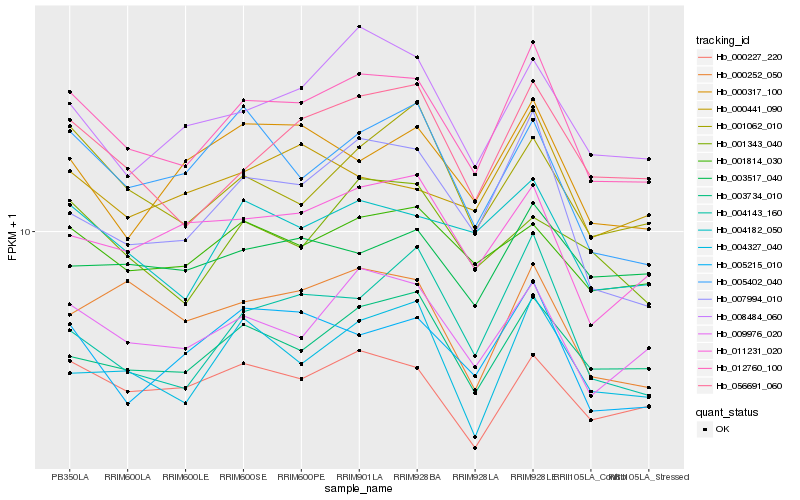
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012760_100 |
0.0 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 2 |
Hb_009976_020 |
0.0536016518 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 3 |
Hb_004182_050 |
0.0560677876 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_056691_060 |
0.0570756469 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007994_010 |
0.0586281599 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003734_010 |
0.0595759808 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 7 |
Hb_005215_010 |
0.061122011 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008484_060 |
0.0637436335 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 9 |
Hb_000227_220 |
0.0650165845 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 10 |
Hb_001814_030 |
0.0651123039 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 11 |
Hb_004327_040 |
0.0655592606 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |
| 12 |
Hb_011231_020 |
0.0666525955 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001062_010 |
0.0687079377 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 14 |
Hb_004143_160 |
0.0696192944 |
- |
- |
SAB, putative [Ricinus communis] |
| 15 |
Hb_000441_090 |
0.0697620556 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 16 |
Hb_000317_100 |
0.0718167188 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 17 |
Hb_005402_040 |
0.0721715143 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 18 |
Hb_001343_040 |
0.0737014566 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 19 |
Hb_003517_040 |
0.0737277882 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000252_050 |
0.0747477464 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |