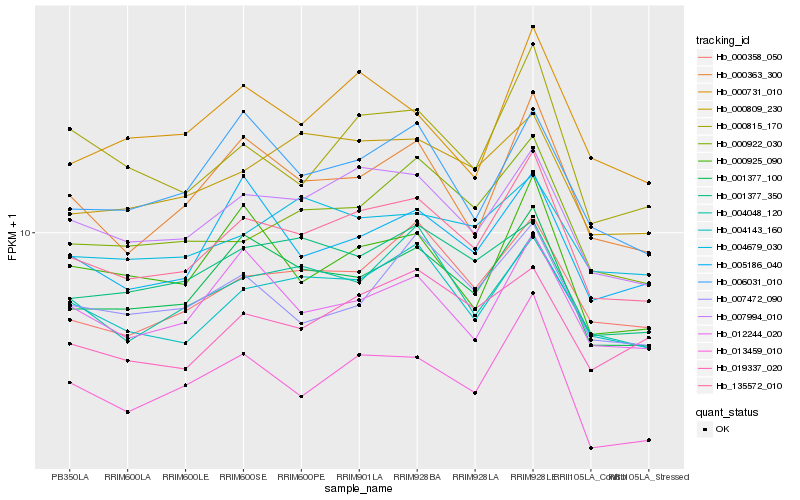
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135572_010 |
0.0 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 2 |
Hb_007994_010 |
0.0522170325 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001377_100 |
0.0574073783 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004143_160 |
0.0607850585 |
- |
- |
SAB, putative [Ricinus communis] |
| 5 |
Hb_000358_050 |
0.065127562 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 6 |
Hb_000363_300 |
0.0680930234 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000922_030 |
0.071319839 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000815_170 |
0.0756102114 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 9 |
Hb_000809_230 |
0.0768088672 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 10 |
Hb_019337_020 |
0.0809365605 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 11 |
Hb_012244_020 |
0.0810667979 |
- |
- |
calpain, putative [Ricinus communis] |
| 12 |
Hb_001377_350 |
0.0820685555 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 13 |
Hb_004048_120 |
0.0820812048 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 14 |
Hb_013459_010 |
0.0827486801 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 15 |
Hb_007472_090 |
0.0829661 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_005186_040 |
0.0831241611 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006031_010 |
0.0849457454 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 18 |
Hb_000925_090 |
0.0850689227 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004679_030 |
0.0857104947 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 20 |
Hb_000731_010 |
0.085962145 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |