| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
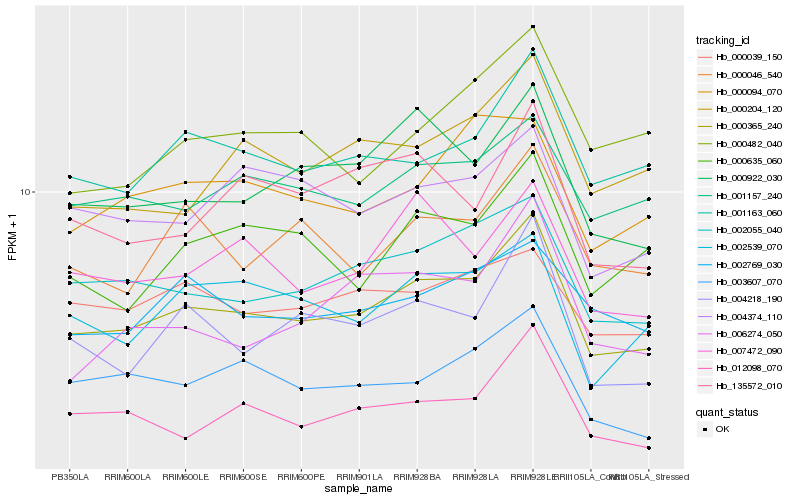
Hb_000365_240 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 2 |
Hb_002055_040 |
0.066301461 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 3 |
Hb_001163_060 |
0.0751530023 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 4 |
Hb_000922_030 |
0.076249851 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001157_240 |
0.0821121672 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 6 |
Hb_002539_070 |
0.0839376887 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 7 |
Hb_004218_190 |
0.0844511014 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000046_540 |
0.0851345949 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 9 |
Hb_007472_090 |
0.08558161 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_000635_060 |
0.0867370284 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 11 |
Hb_000204_120 |
0.0880889801 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 12 |
Hb_002769_030 |
0.0888058395 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_004374_110 |
0.0897257347 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 14 |
Hb_003607_070 |
0.0897321772 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910 [Jatropha curcas] |
| 15 |
Hb_000039_150 |
0.090483214 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000094_070 |
0.0922969302 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_135572_010 |
0.0923167499 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 18 |
Hb_000482_040 |
0.0936569828 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 19 |
Hb_012098_070 |
0.0947572625 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 20 |
Hb_006274_050 |
0.0959332994 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |