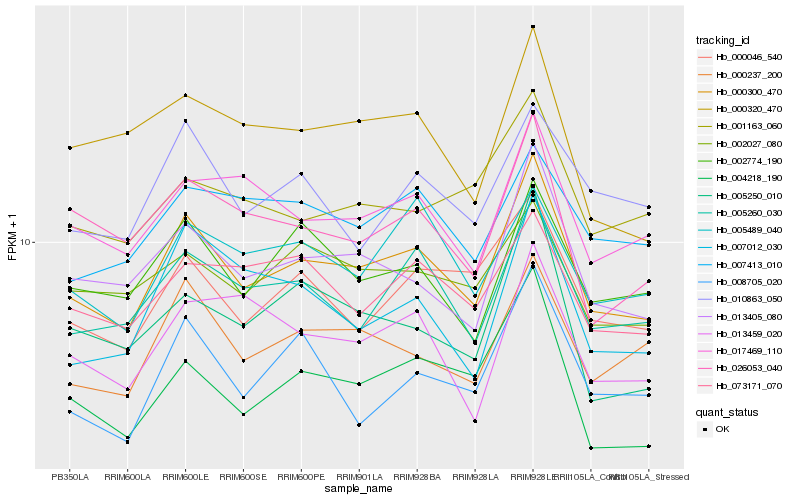
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_470 |
0.0 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 2 |
Hb_013405_080 |
0.0665059203 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 3 |
Hb_005260_030 |
0.0717844389 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 4 |
Hb_000237_200 |
0.0745376323 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 5 |
Hb_002774_190 |
0.0789181929 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 6 |
Hb_000046_540 |
0.0792567722 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 7 |
Hb_004218_190 |
0.0795612942 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |
| 8 |
Hb_017469_110 |
0.0825819492 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 9 |
Hb_026053_040 |
0.086133865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005489_040 |
0.0872069399 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 11 |
Hb_002027_080 |
0.0879171625 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 12 |
Hb_073171_070 |
0.0881949098 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 13 |
Hb_005250_010 |
0.0885474786 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 14 |
Hb_008705_020 |
0.0896328853 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_007012_030 |
0.0900564181 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000320_470 |
0.090391811 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 17 |
Hb_007413_010 |
0.0913892017 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 18 |
Hb_013459_020 |
0.091533341 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 19 |
Hb_010863_050 |
0.0915401412 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 20 |
Hb_001163_060 |
0.0918624323 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |