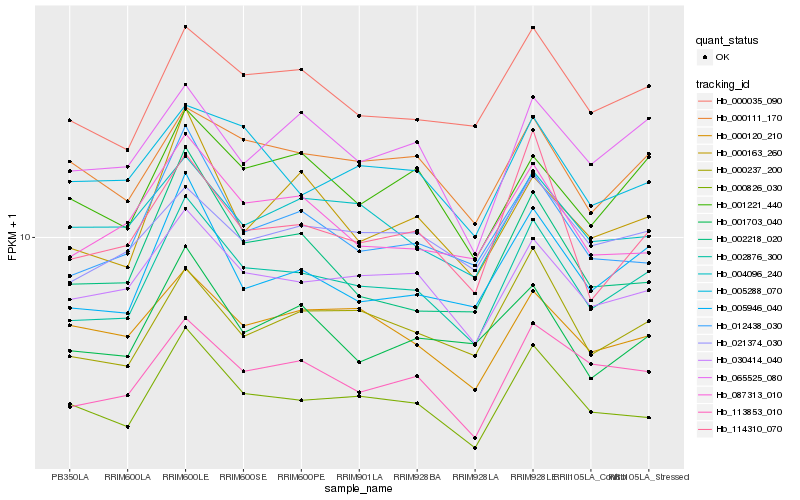
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_300 |
0.0 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_030414_040 |
0.065768787 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000826_030 |
0.0666249898 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 4 |
Hb_000035_090 |
0.067191656 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 5 |
Hb_005946_040 |
0.0677054666 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 6 |
Hb_113853_010 |
0.0700945803 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 7 |
Hb_012438_030 |
0.0740999944 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 8 |
Hb_000237_200 |
0.075061918 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 9 |
Hb_087313_010 |
0.0794944934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001221_440 |
0.0818759728 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_114310_070 |
0.0820919357 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 12 |
Hb_021374_030 |
0.0826744564 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 13 |
Hb_000163_260 |
0.0830315449 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 14 |
Hb_000111_170 |
0.0837415781 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001703_040 |
0.0852165675 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 16 |
Hb_065525_080 |
0.086323435 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 17 |
Hb_004096_240 |
0.0880515701 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 18 |
Hb_005288_070 |
0.0882829076 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000120_210 |
0.0894181917 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 20 |
Hb_002218_020 |
0.0895980075 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |