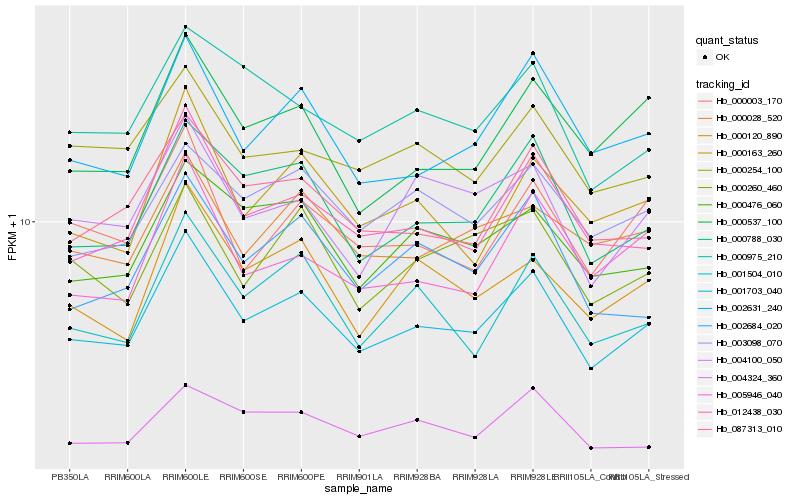
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001703_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000788_030 |
0.0599595867 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 3 |
Hb_012438_030 |
0.0609423665 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 4 |
Hb_002631_240 |
0.0658144702 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 5 |
Hb_087313_010 |
0.0676749124 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001504_010 |
0.0713549329 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 7 |
Hb_000120_890 |
0.0747132675 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 8 |
Hb_000476_060 |
0.0747599085 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_000163_260 |
0.0758261767 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 10 |
Hb_003098_070 |
0.0771194981 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 11 |
Hb_004100_050 |
0.0772691784 |
- |
- |
PREDICTED: mitochondrial-processing peptidase subunit alpha-like [Jatropha curcas] |
| 12 |
Hb_002684_020 |
0.0774805641 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 13 |
Hb_000028_520 |
0.0784783171 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 14 |
Hb_000537_100 |
0.0788031264 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_004324_360 |
0.078824663 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000003_170 |
0.0793027045 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 17 |
Hb_000254_100 |
0.0793208978 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 18 |
Hb_005946_040 |
0.0797866394 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000975_210 |
0.0800593426 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 20 |
Hb_000260_460 |
0.0803555889 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |