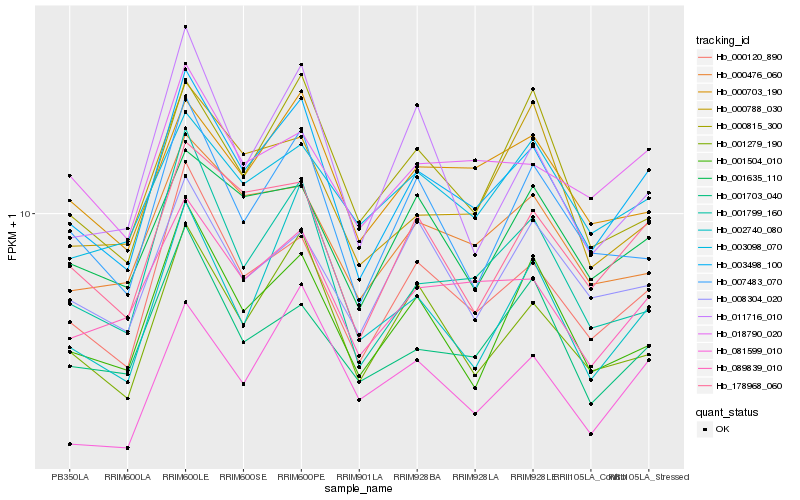
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003498_100 |
0.0 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 2 |
Hb_000120_890 |
0.0474396837 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 3 |
Hb_178968_060 |
0.064709908 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 4 |
Hb_089839_010 |
0.0678324188 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 5 |
Hb_001279_190 |
0.067838744 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000703_190 |
0.0732736212 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001799_160 |
0.0734837999 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_001504_010 |
0.0749152171 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 9 |
Hb_001635_110 |
0.0749364684 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 10 |
Hb_000476_060 |
0.0756125368 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_007483_070 |
0.0756660114 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 12 |
Hb_081599_010 |
0.0794471328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001703_040 |
0.0819312648 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 14 |
Hb_018790_020 |
0.0825182566 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000815_300 |
0.0835861077 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 16 |
Hb_008304_020 |
0.0841268751 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 17 |
Hb_003098_070 |
0.084240307 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 18 |
Hb_002740_080 |
0.0867530749 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 19 |
Hb_000788_030 |
0.0892729098 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 20 |
Hb_011716_010 |
0.0895704645 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |