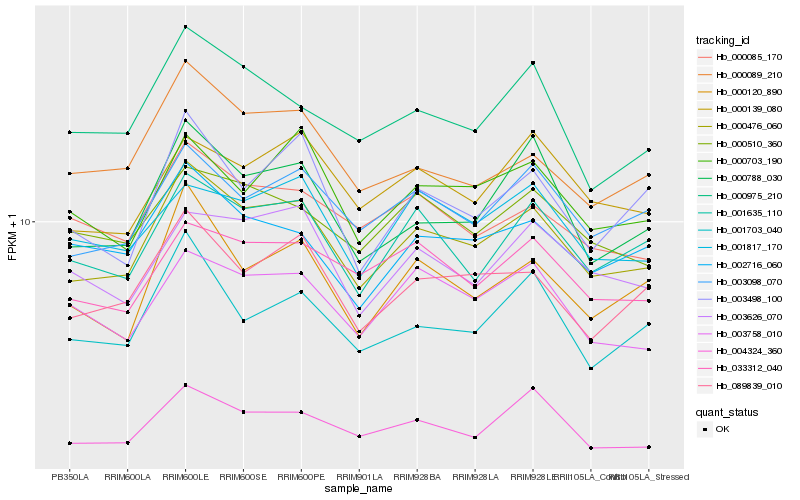
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000476_060 |
0.0 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_003098_070 |
0.0590737322 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 3 |
Hb_089839_010 |
0.0611140052 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 4 |
Hb_001635_110 |
0.0624152458 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 5 |
Hb_000788_030 |
0.0642145579 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 6 |
Hb_004324_360 |
0.0654919558 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000120_890 |
0.0694414077 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 8 |
Hb_000703_190 |
0.0711308881 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000089_210 |
0.0711337879 |
- |
- |
unknown [Medicago truncatula] |
| 10 |
Hb_003626_070 |
0.0713807118 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 11 |
Hb_000139_080 |
0.0714780906 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_033312_040 |
0.0722015747 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000975_210 |
0.0722767419 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 14 |
Hb_000085_170 |
0.0724008017 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002716_060 |
0.072877974 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 16 |
Hb_003758_010 |
0.0730296825 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000510_360 |
0.0732235211 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 18 |
Hb_001817_170 |
0.0746780216 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 19 |
Hb_001703_040 |
0.0747599085 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003498_100 |
0.0756125368 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |