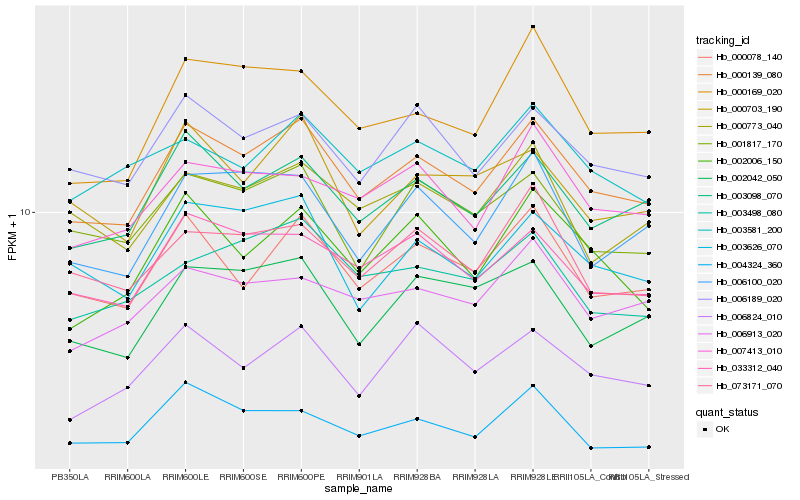
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_080 |
0.0 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003098_070 |
0.0472905079 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 3 |
Hb_033312_040 |
0.0540352996 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000078_140 |
0.0554180329 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 5 |
Hb_006189_020 |
0.0556902888 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 6 |
Hb_003626_070 |
0.0611883971 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 7 |
Hb_000169_020 |
0.0624171231 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 8 |
Hb_001817_170 |
0.0634234489 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 9 |
Hb_006100_020 |
0.0637918026 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 10 |
Hb_073171_070 |
0.0653983494 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 11 |
Hb_006913_020 |
0.0659586421 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007413_010 |
0.0659820375 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 13 |
Hb_000703_190 |
0.0668076646 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002006_150 |
0.0668278567 |
- |
- |
copine, putative [Ricinus communis] |
| 15 |
Hb_002042_050 |
0.067028824 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004324_360 |
0.0674671503 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000773_040 |
0.0676148088 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003581_200 |
0.067990241 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 19 |
Hb_006824_010 |
0.0685884912 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 20 |
Hb_003498_080 |
0.0693344103 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |