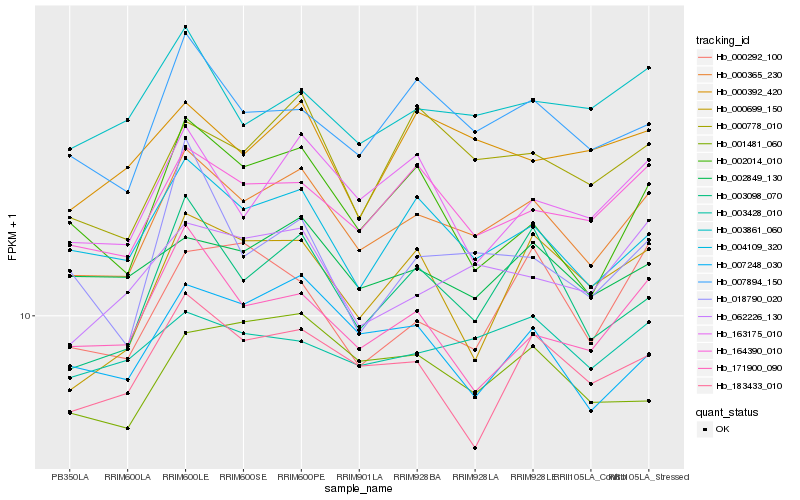
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 2 |
Hb_164390_010 |
0.0498340421 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 3 |
Hb_004109_320 |
0.0501517845 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 4 |
Hb_007894_150 |
0.0539807379 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 5 |
Hb_003098_070 |
0.0561294186 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 6 |
Hb_062226_130 |
0.0570972258 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 7 |
Hb_000778_010 |
0.0581492828 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 8 |
Hb_003861_060 |
0.0602412291 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 9 |
Hb_002849_130 |
0.0606906014 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 10 |
Hb_183433_010 |
0.0611106205 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 11 |
Hb_163175_010 |
0.061884671 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 12 |
Hb_003428_010 |
0.062173657 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 13 |
Hb_007248_030 |
0.0628360399 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 14 |
Hb_171900_090 |
0.0644974255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001481_060 |
0.0662893199 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002014_010 |
0.0666238059 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_000392_420 |
0.066866766 |
- |
- |
PREDICTED: probable aldehyde dehydrogenase isoform X1 [Jatropha curcas] |
| 18 |
Hb_018790_020 |
0.070754955 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 19 |
Hb_000292_100 |
0.0712593213 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 20 |
Hb_000699_150 |
0.0713998904 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |