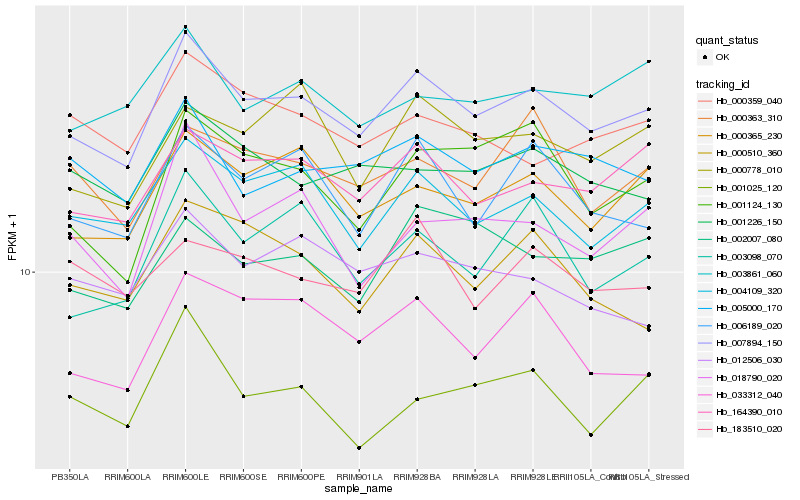
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_150 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 2 |
Hb_004109_320 |
0.0495997199 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 3 |
Hb_164390_010 |
0.0513838038 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 4 |
Hb_000365_230 |
0.0539807379 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 5 |
Hb_006189_020 |
0.0597383393 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 6 |
Hb_005000_170 |
0.0605085095 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 7 |
Hb_001226_150 |
0.0628073881 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 8 |
Hb_033312_040 |
0.0645534508 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003098_070 |
0.0651907757 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 10 |
Hb_001124_130 |
0.0652837293 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 11 |
Hb_003861_060 |
0.0658456368 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 12 |
Hb_018790_020 |
0.0659034785 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 13 |
Hb_002007_080 |
0.0662443583 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 14 |
Hb_000363_310 |
0.0663675906 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 15 |
Hb_000778_010 |
0.067864946 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 16 |
Hb_000510_360 |
0.0698193804 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 17 |
Hb_183510_020 |
0.0699077608 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 18 |
Hb_000359_040 |
0.0702376325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001025_120 |
0.0705988873 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_012506_030 |
0.0708701018 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |