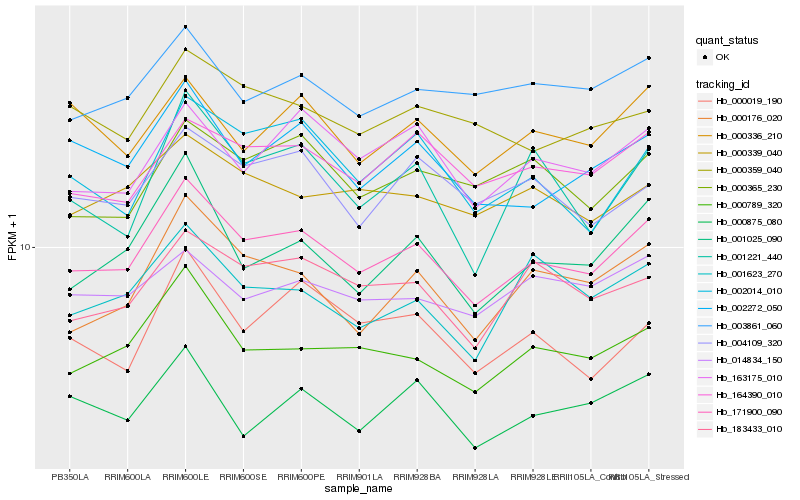
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171900_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000789_320 |
0.0528502084 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_164390_010 |
0.0573627214 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 4 |
Hb_183433_010 |
0.0580166537 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 5 |
Hb_002272_050 |
0.0580596933 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 6 |
Hb_000176_020 |
0.0589447763 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 7 |
Hb_001025_090 |
0.0596363191 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 8 |
Hb_163175_010 |
0.0597855866 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 9 |
Hb_003861_060 |
0.0620427499 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 10 |
Hb_002014_010 |
0.06355348 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_004109_320 |
0.0637578906 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 12 |
Hb_000339_040 |
0.0641640209 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_000365_230 |
0.0644974255 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 14 |
Hb_001623_270 |
0.0654361636 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 15 |
Hb_000336_210 |
0.0662921807 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 16 |
Hb_014834_150 |
0.0674493658 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_000359_040 |
0.06774387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001221_440 |
0.0689214098 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000019_190 |
0.0690357182 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 20 |
Hb_000875_080 |
0.0693756237 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |