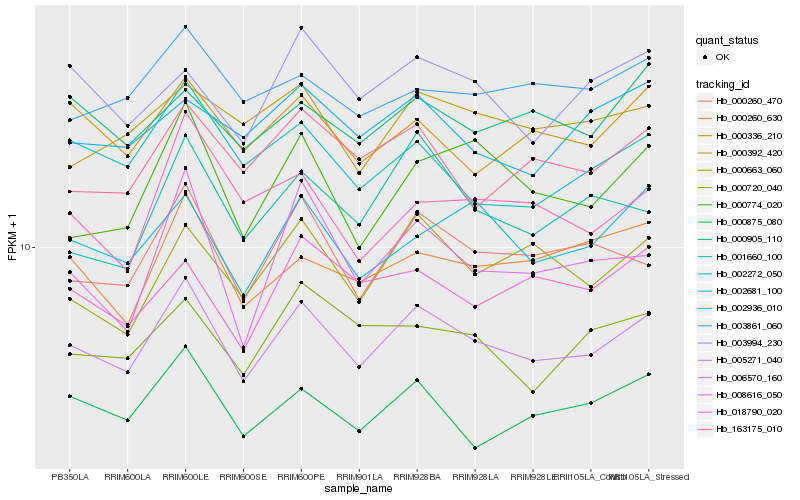
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005271_040 |
0.0 |
- |
- |
PREDICTED: serine racemase [Jatropha curcas] |
| 2 |
Hb_000260_470 |
0.0609174204 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 3 |
Hb_000875_080 |
0.0658290834 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 4 |
Hb_163175_010 |
0.0696442958 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 5 |
Hb_002681_100 |
0.0702579532 |
- |
- |
PREDICTED: glucuronokinase 1 [Jatropha curcas] |
| 6 |
Hb_000663_060 |
0.0709184014 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 7 |
Hb_002272_050 |
0.0717454735 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 8 |
Hb_000336_210 |
0.0719636042 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000774_020 |
0.0731028489 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 10 |
Hb_000260_630 |
0.073229767 |
- |
- |
PREDICTED: altered inheritance rate of mitochondria protein 25 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001660_100 |
0.0750028226 |
- |
- |
PREDICTED: glyoxylate reductase/hydroxypyruvate reductase [Jatropha curcas] |
| 12 |
Hb_000720_040 |
0.0761111483 |
- |
- |
PREDICTED: replication factor C subunit 2 [Jatropha curcas] |
| 13 |
Hb_003861_060 |
0.0764011514 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 14 |
Hb_000392_420 |
0.076643414 |
- |
- |
PREDICTED: probable aldehyde dehydrogenase isoform X1 [Jatropha curcas] |
| 15 |
Hb_006570_160 |
0.0768099357 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 16 |
Hb_003994_230 |
0.0769725555 |
- |
- |
7-dehydrocholesterol reductase, putative [Ricinus communis] |
| 17 |
Hb_008616_050 |
0.0771640972 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 18 |
Hb_000905_110 |
0.0785772577 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 19 |
Hb_002936_010 |
0.0787245813 |
- |
- |
PREDICTED: uncharacterized protein LOC103949110 [Pyrus x bretschneideri] |
| 20 |
Hb_018790_020 |
0.0798572149 |
- |
- |
protein kinase, putative [Ricinus communis] |