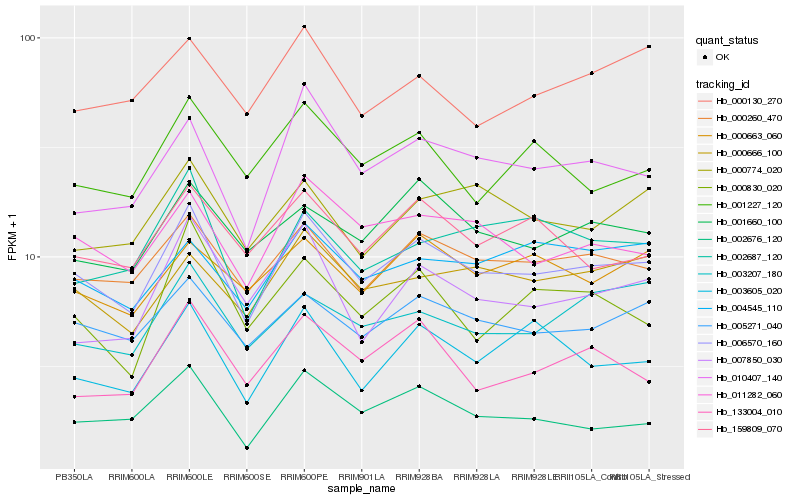
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006570_160 |
0.0 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 2 |
Hb_000260_470 |
0.0694430159 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 3 |
Hb_011282_060 |
0.076244304 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 4 |
Hb_005271_040 |
0.0768099357 |
- |
- |
PREDICTED: serine racemase [Jatropha curcas] |
| 5 |
Hb_010407_140 |
0.0833312931 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 6 |
Hb_002676_120 |
0.0895282572 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 7 |
Hb_000666_100 |
0.0924433561 |
- |
- |
Actin-related protein 2/3 complex subunit 2 isoform 2 [Theobroma cacao] |
| 8 |
Hb_001660_100 |
0.0926179225 |
- |
- |
PREDICTED: glyoxylate reductase/hydroxypyruvate reductase [Jatropha curcas] |
| 9 |
Hb_159809_070 |
0.0935628845 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 10 |
Hb_001227_120 |
0.0946359634 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 11 |
Hb_003605_020 |
0.0965112624 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 12 |
Hb_004545_110 |
0.0999756088 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_000663_060 |
0.1005376045 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 14 |
Hb_000774_020 |
0.1005728863 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 15 |
Hb_000830_020 |
0.1018948738 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 16 |
Hb_003207_180 |
0.1019100672 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002687_120 |
0.1022769628 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_133004_010 |
0.1023163661 |
- |
- |
PREDICTED: cyclin-D3-3 [Jatropha curcas] |
| 19 |
Hb_000130_270 |
0.1028254475 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_007850_030 |
0.1031855344 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |