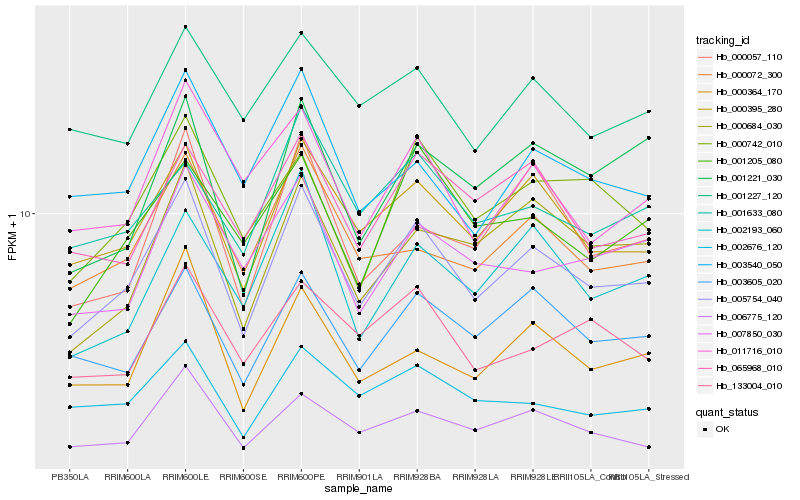
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005754_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000395_280 |
0.0708646219 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 3 |
Hb_003605_020 |
0.0880245377 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 4 |
Hb_002676_120 |
0.0959986227 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 5 |
Hb_006775_120 |
0.0975802018 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 6 |
Hb_000072_300 |
0.0979674151 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 7 |
Hb_000684_030 |
0.0998488096 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 8 |
Hb_011716_010 |
0.1023684577 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 9 |
Hb_000364_170 |
0.103686273 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 10 |
Hb_003540_050 |
0.1037512897 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 11 |
Hb_000742_010 |
0.1038548258 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_065968_010 |
0.1042351505 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000057_110 |
0.1046712019 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 14 |
Hb_002193_060 |
0.1059313744 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 15 |
Hb_007850_030 |
0.1072225118 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 16 |
Hb_001221_030 |
0.108032826 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 17 |
Hb_001633_080 |
0.1081767449 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 18 |
Hb_001227_120 |
0.108769184 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 19 |
Hb_133004_010 |
0.1093905219 |
- |
- |
PREDICTED: cyclin-D3-3 [Jatropha curcas] |
| 20 |
Hb_001205_080 |
0.110083335 |
- |
- |
Protein FAM96B, putative [Ricinus communis] |