| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
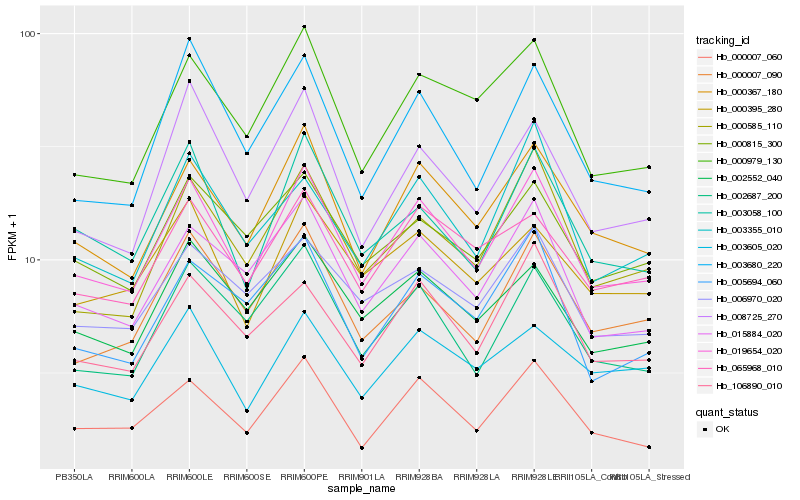
Hb_019654_020 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 2 |
Hb_000367_180 |
0.0525095487 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 3 |
Hb_000007_060 |
0.062831016 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_015884_020 |
0.0674908688 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 5 |
Hb_000979_130 |
0.0717989159 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 6 |
Hb_000585_110 |
0.0731062394 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 7 |
Hb_003058_100 |
0.0745902573 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_106890_010 |
0.0746355356 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_000007_090 |
0.0775865546 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 10 |
Hb_002552_040 |
0.0792831529 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 11 |
Hb_065968_010 |
0.0792840659 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002687_200 |
0.0797341957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003680_220 |
0.0801628785 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 14 |
Hb_000815_300 |
0.0816356954 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 15 |
Hb_006970_020 |
0.0820621034 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 16 |
Hb_000395_280 |
0.0829175018 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 17 |
Hb_003355_010 |
0.0844365264 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 18 |
Hb_008725_270 |
0.08572454 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 19 |
Hb_005694_060 |
0.0857310486 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 20 |
Hb_003605_020 |
0.0858027137 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |