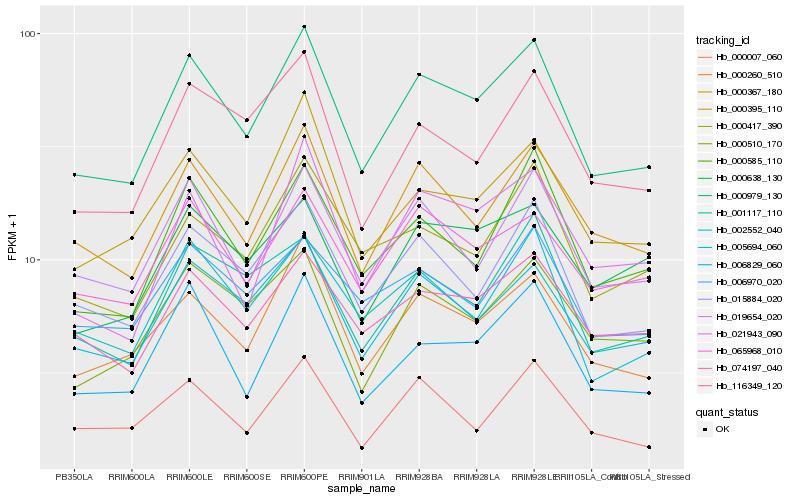
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000979_130 |
0.0 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 2 |
Hb_005694_060 |
0.0659486831 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 3 |
Hb_000260_510 |
0.0665524899 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000367_180 |
0.0675458558 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 5 |
Hb_015884_020 |
0.0687502311 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 6 |
Hb_019654_020 |
0.0717989159 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 7 |
Hb_000395_110 |
0.0776385321 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 8 |
Hb_065968_010 |
0.0793969143 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 9 |
Hb_074197_040 |
0.0799230257 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 10 |
Hb_006829_060 |
0.0799726385 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000585_110 |
0.0816740267 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 12 |
Hb_116349_120 |
0.0843632191 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_000510_170 |
0.0849505549 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 14 |
Hb_001117_110 |
0.0861078845 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 15 |
Hb_000417_390 |
0.0889430866 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 16 |
Hb_002552_040 |
0.0899203859 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 17 |
Hb_021943_090 |
0.0906705119 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 18 |
Hb_006970_020 |
0.0918283319 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 19 |
Hb_000007_060 |
0.0924924556 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000638_130 |
0.0935556137 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |