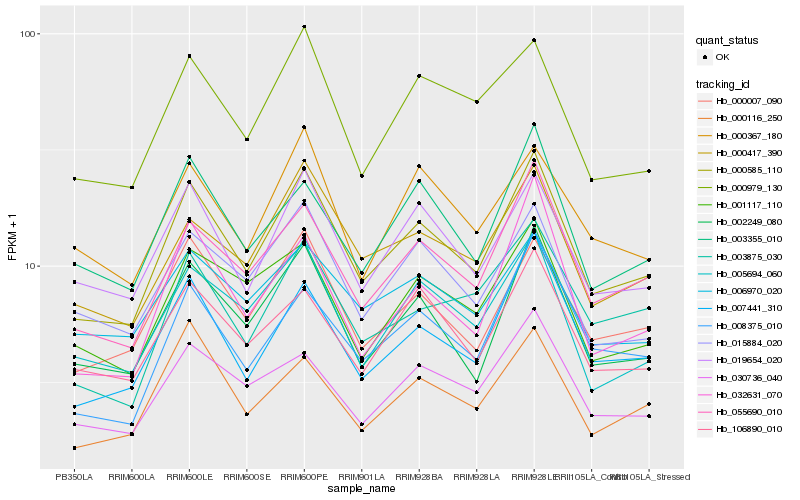
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000585_110 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 2 |
Hb_055690_010 |
0.0594954651 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002249_080 |
0.0649091451 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 4 |
Hb_000007_090 |
0.0656117127 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 5 |
Hb_007441_310 |
0.069059941 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_019654_020 |
0.0731062394 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 7 |
Hb_000417_390 |
0.0759133746 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 8 |
Hb_005694_060 |
0.0778794886 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 9 |
Hb_030736_040 |
0.0785874915 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003355_010 |
0.080979455 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 11 |
Hb_106890_010 |
0.0810264216 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_000979_130 |
0.0816740267 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 13 |
Hb_008375_010 |
0.0818404167 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 14 |
Hb_015884_020 |
0.0835505956 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 15 |
Hb_001117_110 |
0.0835549177 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 16 |
Hb_000116_250 |
0.0857285982 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 17 |
Hb_032631_070 |
0.0871062752 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_003875_030 |
0.0895473675 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 19 |
Hb_006970_020 |
0.0912324046 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 20 |
Hb_000367_180 |
0.0934409853 |
- |
- |
Heparanase-2, putative [Ricinus communis] |