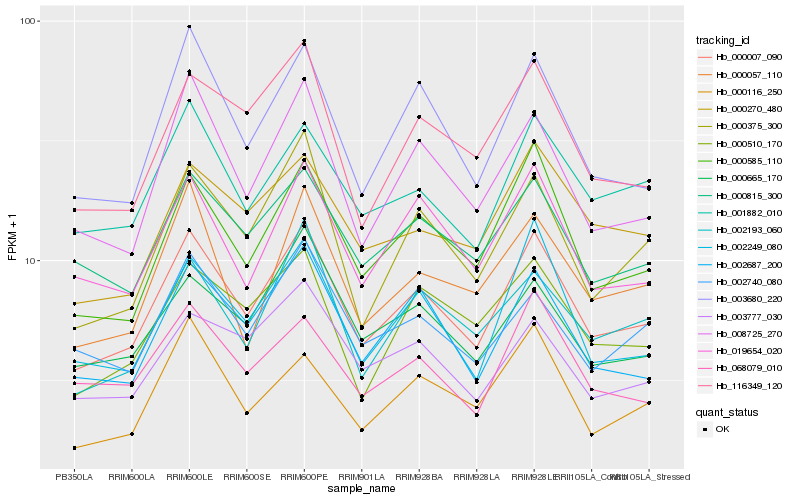
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000007_090 |
0.0 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 2 |
Hb_000585_110 |
0.0656117127 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 3 |
Hb_002687_200 |
0.0718084069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003680_220 |
0.0724438253 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 5 |
Hb_002249_080 |
0.0758872324 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 6 |
Hb_008725_270 |
0.0761656897 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 7 |
Hb_019654_020 |
0.0775865546 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 8 |
Hb_001882_010 |
0.0786749059 |
- |
- |
- |
| 9 |
Hb_000815_300 |
0.0799135003 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 10 |
Hb_000375_300 |
0.0801946858 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 11 |
Hb_116349_120 |
0.0821199486 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_000270_480 |
0.0849121045 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 13 |
Hb_002193_060 |
0.0864940644 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 14 |
Hb_000665_170 |
0.0871833711 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 15 |
Hb_068079_010 |
0.0876345817 |
- |
- |
- |
| 16 |
Hb_000510_170 |
0.0886911478 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 17 |
Hb_000116_250 |
0.0906199018 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 18 |
Hb_000057_110 |
0.090976244 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 19 |
Hb_002740_080 |
0.0913652844 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 20 |
Hb_003777_030 |
0.0918614893 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |