| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
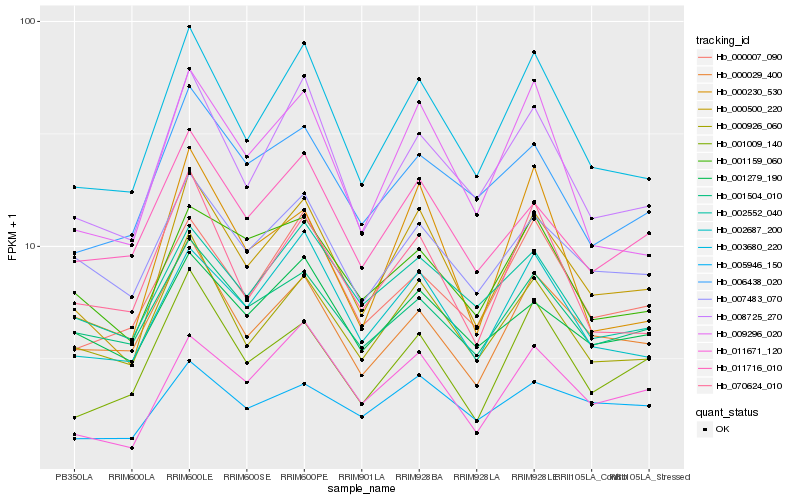
Hb_000500_220 |
0.0 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 2 |
Hb_003680_220 |
0.0704381204 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 3 |
Hb_000926_060 |
0.0797158524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008725_270 |
0.0835516643 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 5 |
Hb_002687_200 |
0.0918361779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011671_120 |
0.0925180499 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |
| 7 |
Hb_009296_020 |
0.0998075563 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_011716_010 |
0.1008582425 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 9 |
Hb_007483_070 |
0.103825281 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 10 |
Hb_070624_010 |
0.10784165 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 11 |
Hb_000029_400 |
0.1080849185 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001279_190 |
0.1097758994 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006438_020 |
0.1103053725 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 14 |
Hb_001009_140 |
0.1103752181 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 15 |
Hb_001159_060 |
0.1119055477 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 16 |
Hb_001504_010 |
0.1121184124 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 17 |
Hb_002552_040 |
0.1130923601 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 18 |
Hb_000007_090 |
0.1136478548 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 19 |
Hb_005946_150 |
0.1147207235 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 20 |
Hb_000230_530 |
0.1151797874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |