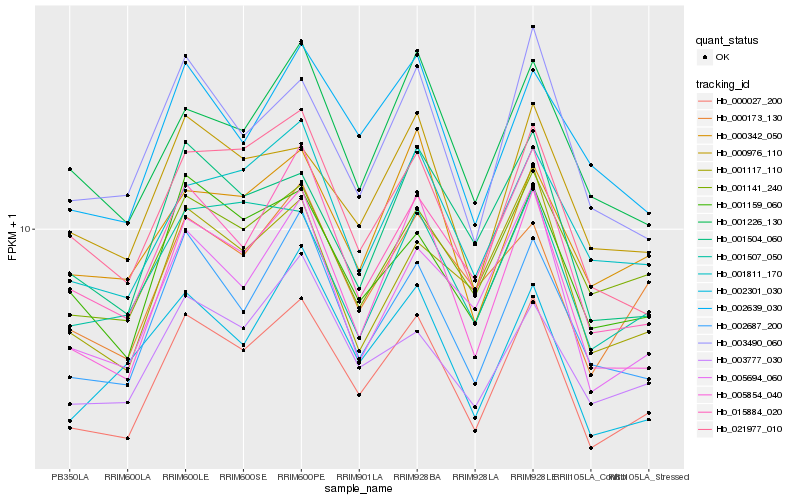
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 2 |
Hb_005854_040 |
0.0559610771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002687_200 |
0.0724268983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_015884_020 |
0.0756271963 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 5 |
Hb_021977_010 |
0.0810351108 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 6 |
Hb_001159_060 |
0.0821257422 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 7 |
Hb_000976_110 |
0.0832601618 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 8 |
Hb_003490_060 |
0.0850616665 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 9 |
Hb_001141_240 |
0.0864083824 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005694_060 |
0.087174948 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 11 |
Hb_001811_170 |
0.0874975428 |
- |
- |
dynamin, putative [Ricinus communis] |
| 12 |
Hb_001507_050 |
0.0892448895 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_001504_060 |
0.0892894262 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 14 |
Hb_002301_030 |
0.0896930205 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 15 |
Hb_003777_030 |
0.0896964494 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 16 |
Hb_000173_130 |
0.0904120035 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002639_030 |
0.0913915902 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 18 |
Hb_001226_130 |
0.0927796869 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 19 |
Hb_001117_110 |
0.0928480568 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 20 |
Hb_000342_050 |
0.0939503685 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |