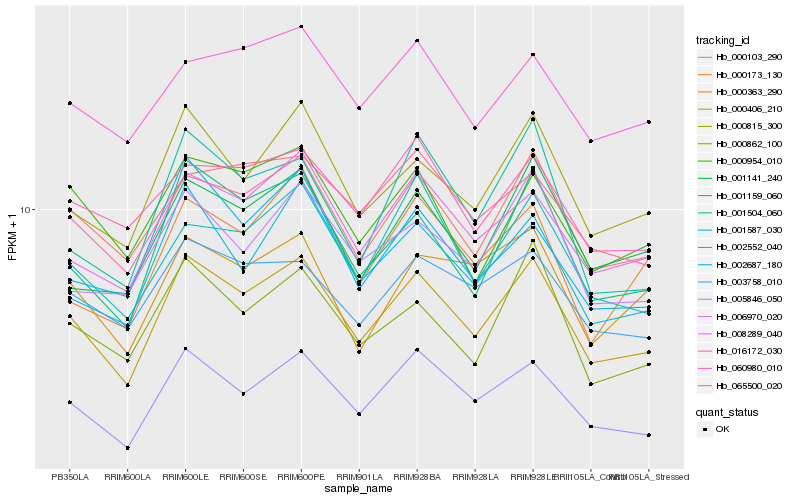
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000406_210 |
0.0 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 2 |
Hb_005846_050 |
0.0554307942 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 3 |
Hb_001159_060 |
0.0589181541 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 4 |
Hb_000815_300 |
0.0615236989 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 5 |
Hb_008289_040 |
0.0645796606 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 6 |
Hb_000103_290 |
0.0686291377 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 7 |
Hb_002552_040 |
0.0727236188 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 8 |
Hb_000862_100 |
0.0743722793 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001587_030 |
0.0753226869 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 10 |
Hb_001141_240 |
0.0768768916 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003758_010 |
0.076900549 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_065500_020 |
0.0774416747 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 13 |
Hb_006970_020 |
0.0793299001 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 14 |
Hb_000954_010 |
0.0795908 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 15 |
Hb_001504_060 |
0.0838750117 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 16 |
Hb_060980_010 |
0.0842006085 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_016172_030 |
0.0845948024 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 18 |
Hb_000363_290 |
0.0865650965 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 19 |
Hb_002687_180 |
0.0865751787 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 20 |
Hb_000173_130 |
0.0866979414 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |