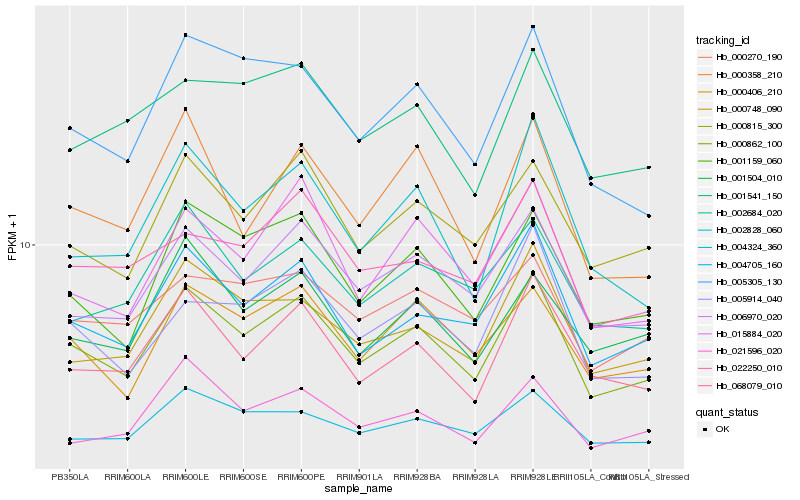
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000862_100 |
0.0 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004705_160 |
0.0645792383 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 3 |
Hb_006970_020 |
0.0664374748 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 4 |
Hb_068079_010 |
0.0687399724 |
- |
- |
- |
| 5 |
Hb_000815_300 |
0.0693362611 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 6 |
Hb_001159_060 |
0.0715938527 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 7 |
Hb_000406_210 |
0.0743722793 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 8 |
Hb_005305_130 |
0.0787436357 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 9 |
Hb_000748_090 |
0.0789394288 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 10 |
Hb_015884_020 |
0.0829465295 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 11 |
Hb_002828_060 |
0.0833085212 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004324_360 |
0.085207498 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002684_020 |
0.0854084432 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 14 |
Hb_005914_040 |
0.0862865313 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 15 |
Hb_022250_010 |
0.0866828783 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 16 |
Hb_000270_190 |
0.0872563711 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 17 |
Hb_000358_210 |
0.0881590213 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 18 |
Hb_001541_150 |
0.0884864015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001504_010 |
0.0889310162 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 20 |
Hb_021596_020 |
0.0893428913 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |