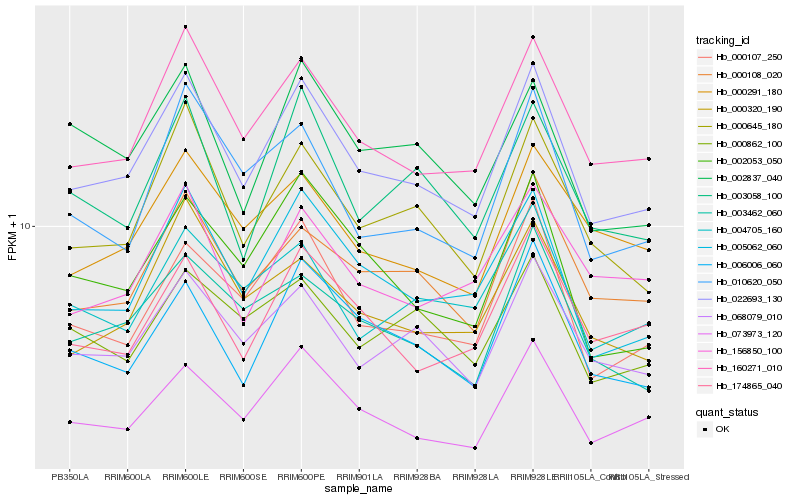
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022693_130 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_160271_010 |
0.057547338 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_005062_060 |
0.0673585456 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 4 |
Hb_068079_010 |
0.0739382662 |
- |
- |
- |
| 5 |
Hb_000645_180 |
0.0762816658 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 6 |
Hb_000108_020 |
0.0768826631 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 7 |
Hb_000107_250 |
0.0807950686 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 8 |
Hb_003462_060 |
0.0890366421 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 9 |
Hb_002053_050 |
0.0892649066 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 10 |
Hb_006006_060 |
0.0895403068 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003058_100 |
0.0906616284 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_174865_040 |
0.0909195733 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_010620_050 |
0.0920142877 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 14 |
Hb_000291_180 |
0.092205678 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 15 |
Hb_004705_160 |
0.0930822947 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 16 |
Hb_073973_120 |
0.0931173766 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_000320_190 |
0.0946400792 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_156850_100 |
0.0959627725 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 19 |
Hb_000862_100 |
0.0976739918 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002837_040 |
0.0976788605 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |