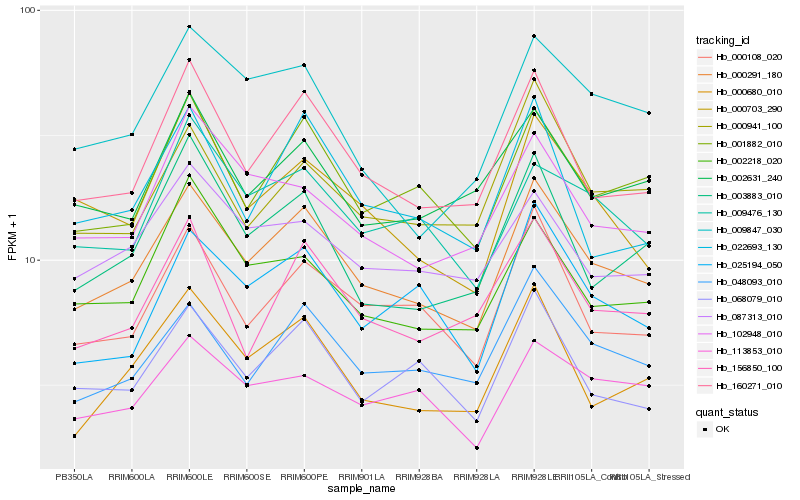
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_180 |
0.0 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 2 |
Hb_160271_010 |
0.0630556668 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001882_010 |
0.0730021814 |
- |
- |
- |
| 4 |
Hb_048093_010 |
0.0766844354 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000108_020 |
0.0803144414 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 6 |
Hb_000680_010 |
0.0867174126 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 7 |
Hb_068079_010 |
0.0867690092 |
- |
- |
- |
| 8 |
Hb_156850_100 |
0.0898522221 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 9 |
Hb_003883_010 |
0.0904607488 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 10 |
Hb_002631_240 |
0.0904830906 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 11 |
Hb_025194_050 |
0.0909701173 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009476_130 |
0.0916256171 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 13 |
Hb_002218_020 |
0.0917917818 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 14 |
Hb_087313_010 |
0.0919209848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_022693_130 |
0.092205678 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_113853_010 |
0.0927620681 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 17 |
Hb_102948_010 |
0.092848987 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000941_100 |
0.0928668587 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 19 |
Hb_009847_030 |
0.0929616645 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 20 |
Hb_000703_290 |
0.093293997 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |