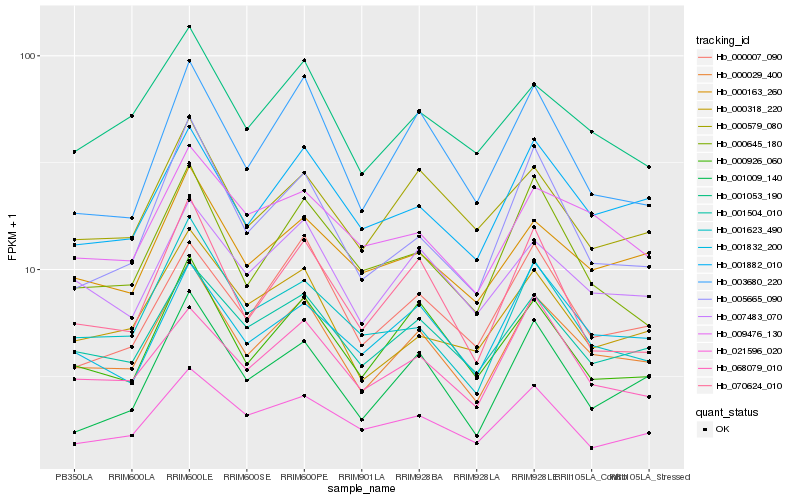
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000029_400 |
0.0 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001504_010 |
0.0693400043 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 3 |
Hb_007483_070 |
0.0769962507 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 4 |
Hb_001882_010 |
0.0782478948 |
- |
- |
- |
| 5 |
Hb_001053_190 |
0.0824728465 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 6 |
Hb_000163_260 |
0.0830514678 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 7 |
Hb_000926_060 |
0.0841076563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000318_220 |
0.0848323982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005665_090 |
0.0870739241 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 10 |
Hb_068079_010 |
0.0889134694 |
- |
- |
- |
| 11 |
Hb_001623_490 |
0.0896971318 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000579_080 |
0.0905479034 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 13 |
Hb_001832_200 |
0.091668041 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003680_220 |
0.0921573814 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 15 |
Hb_009476_130 |
0.0922636966 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 16 |
Hb_070624_010 |
0.0938050229 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 17 |
Hb_021596_020 |
0.0949783007 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 18 |
Hb_000007_090 |
0.0969680279 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 19 |
Hb_000645_180 |
0.0972676895 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 20 |
Hb_001009_140 |
0.0974856415 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |