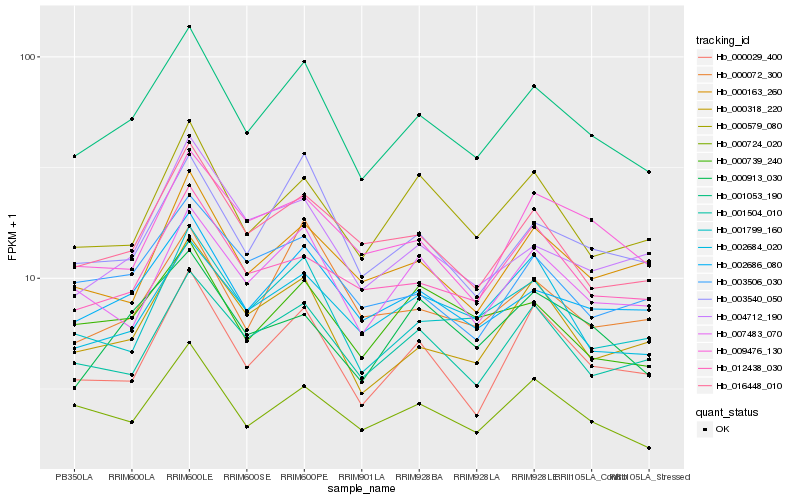
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001053_190 |
0.0 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 2 |
Hb_002686_080 |
0.0651232801 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein ING2-like [Jatropha curcas] |
| 3 |
Hb_003540_050 |
0.0796957854 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 4 |
Hb_000724_020 |
0.0814151734 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 5 |
Hb_000029_400 |
0.0824728465 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000318_220 |
0.0828550005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_016448_010 |
0.0830963778 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 8 |
Hb_001799_160 |
0.0831043662 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_001504_010 |
0.0835721297 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 10 |
Hb_007483_070 |
0.0867013481 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 11 |
Hb_000579_080 |
0.0887908036 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 12 |
Hb_003506_030 |
0.091091348 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_000739_240 |
0.091866903 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_012438_030 |
0.0942945812 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 15 |
Hb_000072_300 |
0.0948211333 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 16 |
Hb_002684_020 |
0.0948578787 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 17 |
Hb_000163_260 |
0.0969327323 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 18 |
Hb_000913_030 |
0.0971697596 |
- |
- |
nuclease, putative [Ricinus communis] |
| 19 |
Hb_009476_130 |
0.0977434755 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 20 |
Hb_004712_190 |
0.1002526465 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |