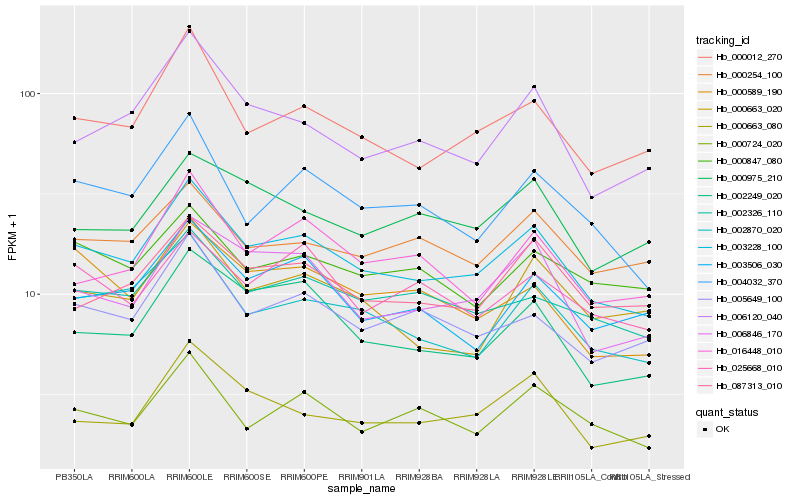
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003228_100 |
0.0 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 2 |
Hb_000012_270 |
0.0707686314 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 3 |
Hb_002870_020 |
0.0722330693 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004032_370 |
0.0779828926 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 5 |
Hb_006846_170 |
0.0783389973 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 6 |
Hb_002326_110 |
0.0784506067 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 7 |
Hb_002249_020 |
0.0802609495 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 8 |
Hb_005649_100 |
0.0810675807 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 9 |
Hb_006120_040 |
0.0846430481 |
- |
- |
PREDICTED: prostatic spermine-binding protein [Jatropha curcas] |
| 10 |
Hb_087313_010 |
0.0851992998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003506_030 |
0.0879855691 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_000724_020 |
0.0889095728 |
transcription factor |
TF Family: PHD |
PREDICTED: probable Histone-lysine N-methyltransferase ATXR5 [Jatropha curcas] |
| 13 |
Hb_000589_190 |
0.0894698565 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_016448_010 |
0.0902795706 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 15 |
Hb_000254_100 |
0.0905885431 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 16 |
Hb_000847_080 |
0.0924269089 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 17 |
Hb_000663_020 |
0.0927148839 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_000663_080 |
0.0929012635 |
transcription factor |
TF Family: HB |
Homeobox protein HAT3.1, putative [Ricinus communis] |
| 19 |
Hb_000975_210 |
0.0932018462 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 20 |
Hb_025668_010 |
0.0937933512 |
- |
- |
unnamed protein product [Coffea canephora] |